8ORS
 
 | | Knockout of GMC-oxidoreductase genes reveals that functional redundancy preserves mimivirus essential functions | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Alempic, J.M, Bisio, H, Villalta, A, Santini, S, Lartigue, A, Schmitt, A, Bugnot, C, Notaro, A, Belmudes, L, Adrait, A, Poirot, O, Ptchelkine, D, De Castro, C, Coute, Y, Abergel, C. | | Deposit date: | 2023-04-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Functional redundancy revealed by the deletion of the mimivirus GMC-oxidoreductase genes.
Microlife, 5, 2024
|
|
8ORH
 
 | | Knockout of GMC-oxidoreductase genes reveals that functional redundancy preserves mimivirus essential functions | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Alempic, J.M, Bisio, H, Villalta, A, Santini, S, Lartigue, A, Schmitt, A, Bugnot, C, Notaro, A, Belmudes, L, Adrait, A, Poirot, O, Ptchelkine, D, De Castro, C, Coute, Y, Abergel, C. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Functional redundancy revealed by the deletion of the mimivirus GMC-oxidoreductase genes.
Microlife, 5, 2024
|
|
1CF3
 
 | | GLUCOSE OXIDASE FROM APERGILLUS NIGER | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (GLUCOSE OXIDASE), ... | | Authors: | Hecht, H.J, Kalisz, H. | | Deposit date: | 1999-03-23 | | Release date: | 1999-03-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.8 and 1.9 A resolution structures of the Penicillium amagasakiense and Aspergillus niger glucose oxidases as a basis for modelling substrate complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
7VZP
 
 | | FAD-dpendent Glucose Dehydrogenase from Aspergillus oryzae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GMC oxidoreductase, PENTAETHYLENE GLYCOL | | Authors: | Nakajima, Y. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational change of catalytic residue in reduced enzyme of FAD-dependent Glucose Dehydrogenase at pH6.5
To Be Published
|
|
3T37
 
 | | Crystal structure of pyridoxine 4-oxidase from Mesorbium loti | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable dehydrogenase | | Authors: | Mugo, A.N, Kobayashi, J, Mikami, B, Ohnishi, K, Yagi, T. | | Deposit date: | 2011-07-25 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Structure biology and crystallization communication
TO BE PUBLISHED
|
|
2GEW
 
 | |
3NNE
 
 | | Crystal structure of choline oxidase S101A mutant | | Descriptor: | ACETATE ION, Choline oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Wang, Y.-F, Finnegan, S, Yuan, H, Orville, A.M, Weber, I.T, Gadda, G. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural and kinetic studies on the Ser101Ala variant of choline oxidase: Catalysis by compromise.
Arch.Biochem.Biophys., 501, 2010
|
|
3RED
 
 | | 3.0 A structure of the Prunus mume hydroxynitrile lyase isozyme-1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Hydroxynitrile lyase | | Authors: | Cielo, C.B.C, Yamane, T, Asano, Y, Watanabe, N, Suzuki, A, Fukuta, Y. | | Deposit date: | 2011-04-04 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Crystal Structure of a native FAD-dependent Hydroxynitrile Lyase derived from the Japanese apricot, Prunus mume
To be Published
|
|
7VZS
 
 | | FAD-dpendent Glucose Dehydrogenase complexed with an inhibitor at pH7.56 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-glucal, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nakajima, Y. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational change of catalytic residue in reduced enzyme of FAD-dependent Glucose Dehydrogenase at pH6.5
To Be Published
|
|
7W2J
 
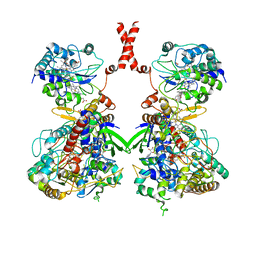 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Makino, F, Miyata, T, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2021-11-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry
Acs Catalysis, 13, 2023
|
|
1B4V
 
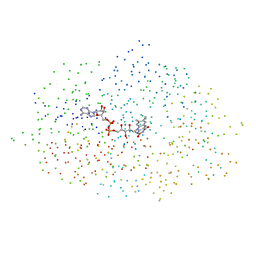 | | CHOLESTEROL OXIDASE FROM STREPTOMYCES | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (CHOLESTEROL OXIDASE) | | Authors: | Vrielink, A, Yue, Q.K. | | Deposit date: | 1998-12-30 | | Release date: | 1999-01-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure determination of cholesterol oxidase from Streptomyces and structural characterization of key active site mutants.
Biochemistry, 38, 1999
|
|
3Q9T
 
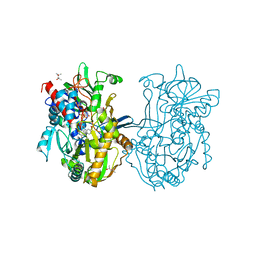 | | Crystal structure analysis of formate oxidase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Doubayashi, D, Ootake, T, Maeda, Y, Oki, M, Tokunaga, Y, Sakurai, A, Nagaosa, Y, Mikami, B, Uchida, H. | | Deposit date: | 2011-01-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Formate oxidase, an enzyme of the glucose-methanol-choline oxidoreductase family, has a His-Arg pair and 8-formyl-FAD at the catalytic site.
Biosci.Biotechnol.Biochem., 75, 2011
|
|
7QFD
 
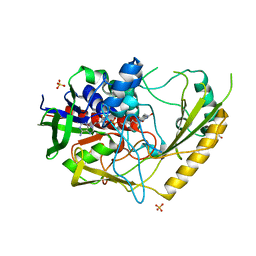 | | Crystal structure of a bacterial pyranose 2-oxidase complex with D-glucose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, GMC oxidoreductase family protein, ... | | Authors: | Borges, P.T, Frazao, T, Taborda, A, Frazao, C, Martins, L.O. | | Deposit date: | 2021-12-05 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanistic insights into glycoside 3-oxidases involved in C-glycoside metabolism in soil microorganisms.
Nat Commun, 14, 2023
|
|
7QF8
 
 | | Crystal structure of a bacterial pyranose 2-oxidase from Pseudoarthrobacter siccitolerans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, GMC oxidoreductase family protein, ... | | Authors: | Borges, P.T, Frazao, T, Taborda, A, Frazao, C, Martins, L.O. | | Deposit date: | 2021-12-04 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Mechanistic insights into glycoside 3-oxidases involved in C-glycoside metabolism in soil microorganisms.
Nat Commun, 14, 2023
|
|
7QVA
 
 | | Crystal structure of a bacterial pyranose 2-oxidase in complex with mangiferin | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GMC oxidoreductase family protein, Mangiferin, ... | | Authors: | Borges, P.T, Frazao, T, Taborda, T, Brissos, V, Frazao, C, Martins, L.O. | | Deposit date: | 2022-01-20 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanistic insights into glycoside 3-oxidases involved in C-glycoside metabolism in soil microorganisms.
Nat Commun, 14, 2023
|
|
7R34
 
 | | Difference-refined structure of fatty acid photodecarboxylase 900 ps following 400-nm laser irradiation of the dark-state determined by SFX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R35
 
 | | Difference-refined structure of fatty acid photodecarboxylase 300 ns following 400-nm laser irradiation of the dark-state determined by SFX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, W. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R33
 
 | | Difference-refined structure of fatty acid photodecarboxylase 20 ps following 400-nm laser irradiation of the dark-state determined by SFX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R36
 
 | | Difference-refined structure of fatty acid photodecarboxylase 2 microsecond following 400-nm laser irradiation of the dark-state determined by SFX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6XUU
 
 | | Crystallographic structure of oligosaccharide dehydrogenase from Pycnoporus cinnabarinus, glucose-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cerutti, G, Savino, C, Montemiglio, L.C, Vallone, B, Sciara, G. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure and functional characterization of an oligosaccharide dehydrogenase from Pycnoporus cinnabarinus provides insights into fungal breakdown of lignocellulose.
Biotechnol Biofuels, 14, 2021
|
|
6XUT
 
 | | Crystallographic structure of oligosaccharide dehydrogenase from Pycnoporus cinnabarinus, ligand-free form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cerutti, G, Savino, C, Montemiglio, L.C, Sciara, G, Vallone, B. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and functional characterization of an oligosaccharide dehydrogenase from Pycnoporus cinnabarinus provides insights into fungal breakdown of lignocellulose.
Biotechnol Biofuels, 14, 2021
|
|
6YRV
 
 | | Crystal structure of FAP after illumination at 100K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRU
 
 | | Crystal structure of FAP in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YS2
 
 | | Crystal structure of FAP R451A in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRX
 
 | | Low-dose crystal structure of FAP at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
