6LUJ
 
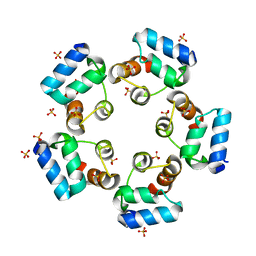 | | Crystal structure of the SAMD1 SAM domain | | Descriptor: | Atherin, SULFATE ION | | Authors: | Cao, Y, Zhou, Y, Wang, Z. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | The SAM domain-containing protein 1 (SAMD1) acts as a repressive chromatin regulator at unmethylated CpG islands.
Sci Adv, 7, 2021
|
|
8VOB
 
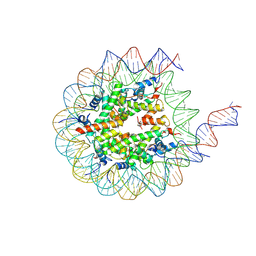 | | H3K36me3-modified nucleosome bound to PRC2_AJ1-450 | | Descriptor: | DNA (157-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-14 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8VNZ
 
 | |
8VO0
 
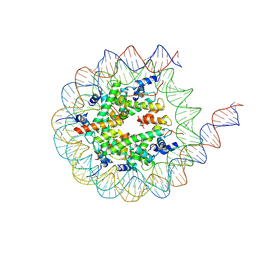 | |
8VNV
 
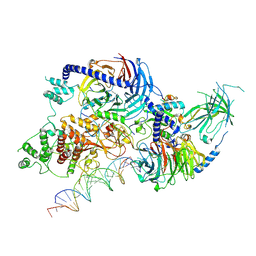 | |
3LPY
 
 | | Crystal structure of the RRM domain of CyP33 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Peptidyl-prolyl cis-trans isomerase E, SULFATE ION | | Authors: | Wang, Z, Patel, D.J. | | Deposit date: | 2010-02-07 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression.
Cell(Cambridge,Mass.), 141, 2010
|
|
3N9M
 
 | | ceKDM7A from C.elegans, alone | | Descriptor: | FE (II) ION, Putative uncharacterized protein, ZINC ION | | Authors: | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | Deposit date: | 2010-05-31 | | Release date: | 2010-06-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
5WXG
 
 | | Structure of TAF PHD finger domain binds to H3(1-15)K4ac | | Descriptor: | Histone H3K4ac, MAGNESIUM ION, Transcription initiation factor TFIID subunit 3, ... | | Authors: | Zhao, S, Li, H. | | Deposit date: | 2017-01-07 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XVW
 
 | |
5Y21
 
 | |
5Y53
 
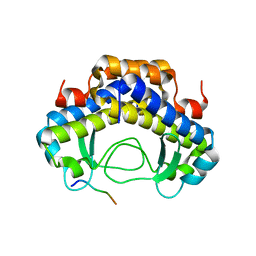 | |
5YVX
 
 | | Crystal structure of SDG8 CW domain in complex with H3K4me1 peptide | | Descriptor: | H3K4me1, Histone-lysine N-methyltransferase ASHH2, ZINC ION | | Authors: | Liu, Y, Huang, Y. | | Deposit date: | 2017-11-27 | | Release date: | 2018-03-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Uncovering the mechanistic basis for specific recognition of monomethylated H3K4 by the CW domain ofArabidopsishistone methyltransferase SDG8.
J. Biol. Chem., 293, 2018
|
|
7ZEY
 
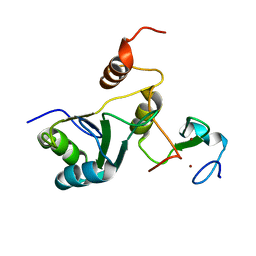 | | Complex Cyp33-RRM : MLL1-PHD3 | | Descriptor: | MLL cleavage product N320, Peptidyl-prolyl cis-trans isomerase E, ZINC ION | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
2NS2
 
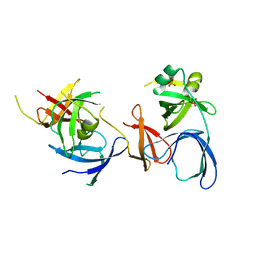 | | Crystal Structure of Spindlin1 | | Descriptor: | PHOSPHATE ION, Spindlin-1 | | Authors: | Zhao, Q, Qin, L, Jiang, F, Wu, B, Yue, W, Xu, F, Rong, Z, Yuan, H, Xie, X, Gao, Y, Bai, C, Bartlam, M. | | Deposit date: | 2006-11-02 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human spindlin1. Tandem tudor-like domains for cell cycle regulation
J.Biol.Chem., 282, 2007
|
|
7CFP
 
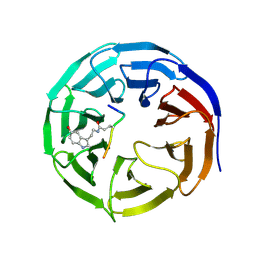 | |
7E9M
 
 | | Crystal Structure of Spindlin1 bound to SPINDOC Docpep3 | | Descriptor: | Peptide from Spindlin interactor and repressor of chromatin-binding protein, Spindlin-1 | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2021-03-04 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for SPINDOC-Spindlin1 engagement and its role in transcriptional inhibition
to be published
|
|
8VLD
 
 | |
8VLH
 
 | |
5D6Y
 
 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A complexed with histone H3K23me3 | | Descriptor: | Lysine-specific demethylase 4A, peptide H3K23me3 (19-28) | | Authors: | Wang, F, Su, Z, Miller, M.D, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2016-02-10 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
6G0L
 
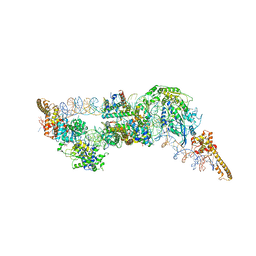 | | Structure of two molecules of the chromatin remodelling enzyme Chd1 bound to a nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Sundaramoorthy, R, Owen-hughes, T, Norman, D.G, Hughes, A. | | Deposit date: | 2018-03-19 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome.
Elife, 7, 2018
|
|
5O9G
 
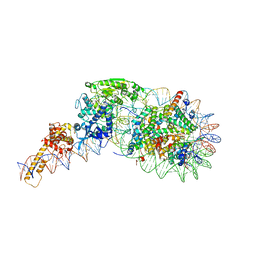 | | Structure of nucleosome-Chd1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Farnung, L, Vos, S.M, Wigge, C, Cramer, P. | | Deposit date: | 2017-06-19 | | Release date: | 2017-10-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Nucleosome-Chd1 structure and implications for chromatin remodelling.
Nature, 550, 2017
|
|
7K7T
 
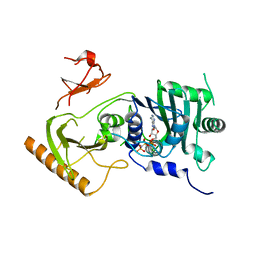 | | Crystal structure of human MORC4 ATPase-CW in complex with AMPPNP | | Descriptor: | Isoform 3 of MORC family CW-type zinc finger protein 4, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Klein, B.J, Tencer, A.H, Kutateladze, T.G. | | Deposit date: | 2020-09-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Molecular mechanism of the MORC4 ATPase activation.
Nat Commun, 11, 2020
|
|
4UXN
 
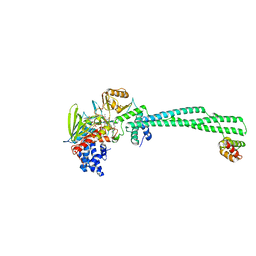 | | LSD1(KDM1A)-CoREST in complex with Z-Pro derivative of MC2580 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, ... | | Authors: | Rodriguez, V, Valente, S, Stazi, G, Lucidi, A, Mercurio, C, Vianello, P, Ciossani, G, Mattevi, A, Botrugno, O.A, Dessanti, P, Minucci, S, Varasi, M, Mai, A. | | Deposit date: | 2014-08-27 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Pyrrole- and Indole-Containing Tranylcypromine Derivatives as Novel Lysine-Specific Demethylase 1 Inhibitors Active on Cancer Cells
Chemmedchem, 6, 2015
|
|
3JBX
 
 | | Cryo-electron microscopy structure of RAG Signal End Complex (C2 symmetry) | | Descriptor: | 5'-D(*CP*AP*CP*AP*GP*TP*GP*CP*TP*AP*CP*AP*GP*AP*C)-3', 5'-D(*GP*CP*GP*AP*TP*GP*GP*TP*TP*AP*AP*CP*CP*A)-3', 5'-D(P*GP*TP*CP*TP*GP*TP*AP*GP*CP*AP*CP*TP*GP*TP*G)-3', ... | | Authors: | Ru, H, Chambers, M.G, Fu, T.-M, Tong, A.B, Liao, M, Wu, H. | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures.
Cell(Cambridge,Mass.), 163, 2015
|
|
7NKX
 
 | | RNA polymerase II-Spt4/5-nucleosome-Chd1 structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromatin elongation factor SPT4, ... | | Authors: | Farnung, L, Ochmann, M, Engeholm, M, Cramer, P. | | Deposit date: | 2021-02-19 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome transcription mediated by Chd1 and FACT.
Nat.Struct.Mol.Biol., 28, 2021
|
|
