8K0R
 
 | |
8K15
 
 | |
1B3S
 
 | |
1B2S
 
 | |
8K0S
 
 | |
8K0Q
 
 | |
8K0P
 
 | |
7BJS
 
 | | Crystal structure of Khc/atypical Tm1 complex | | Descriptor: | Kinesin heavy chain, SD21996p | | Authors: | Dimitrova-Paternoga, L, Jagtap, P.K.A, Ephrussi, A, Hennig, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Molecular basis of mRNA transport by a kinesin-1-atypical tropomyosin complex.
Genes Dev., 35, 2021
|
|
7BJN
 
 | | Crystal structure of atypical Tm1 (Tm1-I/C), residues 270-334 | | Descriptor: | SD21996p | | Authors: | Dimitrova-Paternoga, L, Jagtap, P.K.A, Ephrussi, A, Hennig, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of mRNA transport by a kinesin-1-atypical tropomyosin complex.
Genes Dev., 35, 2021
|
|
1MRE
 
 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), ... | | Authors: | Pokkuluri, P.R, Cygler, M. | | Deposit date: | 1994-06-13 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
6FMB
 
 | |
1B27
 
 | |
6IB8
 
 | | Structure of a complex of SuhB and NusA AR2 domain | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, Inositol-1-monophosphatase, ... | | Authors: | Huang, Y.H, Loll, B, Wahl, M.C. | | Deposit date: | 2018-11-29 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Structural basis for the function of SuhB as a transcription factor in ribosomal RNA synthesis.
Nucleic Acids Res., 47, 2019
|
|
5AN4
 
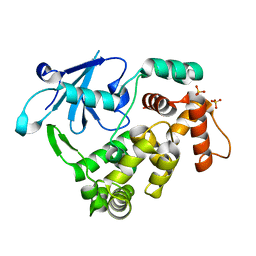 | | Crystal structure of the human 8-oxoguanine glycosylase (OGG1) processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | N-GLYCOSYLASE/DNA LYASE, SULFATE ION | | Authors: | Zander, U, Ytre-Arne, M, Dalhus, B, Hoffmann, G, Cornaciu, I, Cipriani, F, Marquez, J.A. | | Deposit date: | 2015-09-04 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Automated Harvesting and Processing of Protein Crystals Through Laser Photoablation.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
1MRC
 
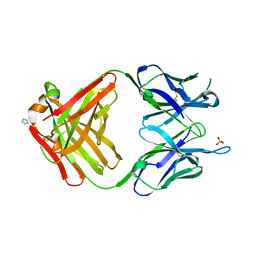 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | Descriptor: | IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), IMIDAZOLE, ... | | Authors: | Pokkuluri, P.R, Cygler, M. | | Deposit date: | 1994-06-13 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
1MRD
 
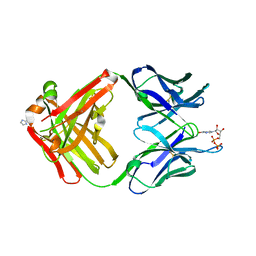 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | Descriptor: | IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), IMIDAZOLE, ... | | Authors: | Pokkuluri, P.R, Cygler, M. | | Deposit date: | 1994-06-13 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
1LN4
 
 | |
1B2U
 
 | |
3LPH
 
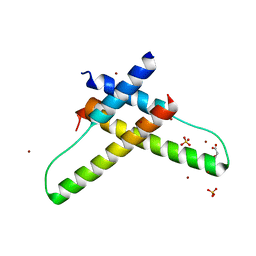 | | Crystal structure of the HIV-1 Rev dimer | | Descriptor: | BROMIDE ION, MALONATE ION, Protein Rev, ... | | Authors: | Daugherty, M.D. | | Deposit date: | 2010-02-05 | | Release date: | 2010-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for cooperative RNA binding and export complex assembly by HIV Rev.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XSF
 
 | |
5W1T
 
 | |
4KB7
 
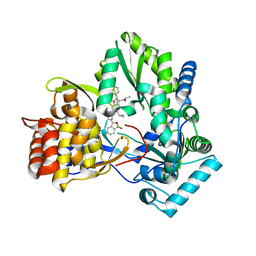 | | HCV NS5B GT1B N316Y with CMPD 32 | | Descriptor: | 5-cyclopropyl-2-(4-fluorophenyl)-6-[{2-[(3R)-1-hydroxy-1,3-dihydro-2,1-benzoxaborol-3-yl]ethyl}(methylsulfonyl)amino]-N-methyl-1-benzofuran-3-carboxamide, HCV Polymerase | | Authors: | Williams, S.P, Kahler, K.M, Shotwell, J.B. | | Deposit date: | 2013-04-23 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of a Potent Boronic Acid Derived Inhibitor of the HCV RNA-Dependent RNA Polymerase.
J.Med.Chem., 57, 2014
|
|
6UF1
 
 | | Pistol ribozyme transition-state analog vanadate | | Descriptor: | MANGANESE (II) ION, RNA (5'-R(*UP*CP*UP*GP*CP*UP*CP*UP*CP*(GVA)*UP*CP*CP*AP*A)-3'), RNA (50-MER) | | Authors: | Teplova, M, Falschlunger, C, Krasheninina, O, Patel, D.J, Micura, R. | | Deposit date: | 2019-09-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crucial Roles of Two Hydrated Mg2+Ions in Reaction Catalysis of the Pistol Ribozyme.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8T66
 
 | | cA6 bound Cam1 | | Descriptor: | Cam1, RNA (5'-R(P*AP*AP*AP*AP*A)-3') | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
8T65
 
 | | cA4 bound Cam1 | | Descriptor: | Cam1, RNA (5'-R(P*AP*AP*AP*A)-3') | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
