8U9E
 
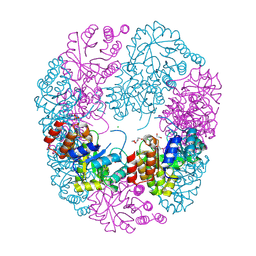 | | Crystal Structure of Staphylococcus aureus Pdx1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Barra, A.L.C, Nascimento, A.S. | | Deposit date: | 2023-09-19 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structure and dynamics of the staphylococcal pyridoxal 5-phosphate synthase complex reveal transient interactions at the enzyme interface.
J.Biol.Chem., 300, 2024
|
|
5H21
 
 | | Trimethoxy-ring inhibitor in complex with the first bromodomain of BRD4 | | Descriptor: | 3,4,5-trimethoxy-~{N}-(2-thiophen-2-ylethyl)benzamide, Bromodomain-containing protein 4 | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Discovery of novel trimethoxy-ring BRD4 bromodomain inhibitors: AlphaScreen assay, crystallography and cell-based assay.
Medchemcomm, 8, 2017
|
|
7NN1
 
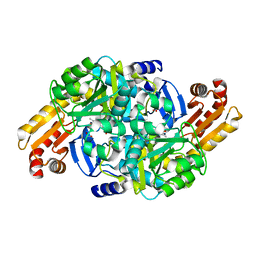 | | Crystal structure of Mycobacterium tuberculosis ArgD with prosthetic group pyridoxal 5'-phosphate | | Descriptor: | Acetylornithine aminotransferase, NITRATE ION | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
8RQJ
 
 | |
7N7I
 
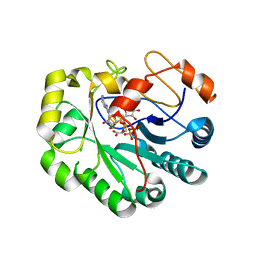 | | X-ray crystal structure of Viperin-like enzyme from Trichoderma virens | | Descriptor: | IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Grove, T.L, Almo, S.C, Bonanno, J.B, Lachowicz, J.C, Gizzi, A.G. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural Insight into the Substrate Scope of Viperin and Viperin-like Enzymes from Three Domains of Life.
Biochemistry, 60, 2021
|
|
8OK6
 
 | | Variant Surface Glycoprotein VSG11 two monomers | | Descriptor: | SULFATE ION, Variant surface glycoprotein, alpha-D-glucopyranose, ... | | Authors: | Gkeka, A, Vlachou, E.P, Zeelen, J.P, Stebbins, C.E. | | Deposit date: | 2023-03-27 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A structural classification of the variant surface glycoproteins of the African trypanosomey.
Plos Negl Trop Dis, 17, 2023
|
|
5CFM
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in complex with 3', 3' cGAMP, c[G(3', 5')pA(3', 5')p] | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, CITRATE ANION, Stimulator of Interferon Genes | | Authors: | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
7VIQ
 
 | | Crystal structure of Au(50EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
5U0M
 
 | | Fatty aldehyde dehydrogenase from Marinobacter aquaeolei VT8 and cofactor complex | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, N-succinylglutamate 5-semialdehyde dehydrogenase, ... | | Authors: | Shi, K, Mulliner, K, Barney, B.M, Aihara, H. | | Deposit date: | 2016-11-24 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.075 Å) | | Cite: | Five Fatty Aldehyde Dehydrogenase Enzymes from Marinobacter and Acinetobacter spp. and Structural Insights into the Aldehyde Binding Pocket.
Appl. Environ. Microbiol., 83, 2017
|
|
8RFT
 
 | | Low pH (5.5) nitrite-bound MSOX movie series dataset 17 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [10.37 MGy] - water ligand + decarboxylated AspCAT | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
7VIS
 
 | | Crystal structure of Au(200EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
5U14
 
 | | E. coli dihydropteroate synthase complexed with an 8-mercaptoguanine derivative: 4-{2-[(2-amino-6-oxo-6,9-dihydro-1H-purin-8-yl)sulfanyl]ethyl}benzene-1-sulfonamide | | Descriptor: | 4-{2-[(2-amino-6-oxo-6,9-dihydro-1H-purin-8-yl)sulfanyl]ethyl}benzene-1-sulfonamide, Dihydropteroate synthase | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2016-11-27 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | 8-Mercaptoguanine Derivatives as Inhibitors of Dihydropteroate Synthase.
Chemistry, 24, 2018
|
|
7NM0
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 1-(2,6-dihydroxyphenyl)ethan-1-one. | | Descriptor: | 1,2-ETHANEDIOL, 1-[2,6-bis(oxidanyl)phenyl]ethanone, Acetylglutamate kinase, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
8RGD
 
 | | High pH (8.0) nitrite-bound MSOX movie series dataset 15 of the copper nitrite reductase from Bradyrhizobium sp. ORS375 (two-domain) [10.2 MGy] | | Descriptor: | CARBON DIOXIDE, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
8OJO
 
 | | Galectin-3 in complex with 2,6-Anhydro-5-S-(beta-D-galactopyranosyl)-5-thio-D-altritol | | Descriptor: | 1-deoxy-alpha-D-mannopyranose, 1-thio-beta-D-galactopyranose, Galectin-3, ... | | Authors: | Tsagkarakou, A.S, Leonidas, D.D. | | Deposit date: | 2023-03-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Strong Binding of C -Glycosylic1,2-Thiodisaccharides to Galectin-3─Enthalpy-Driven Affinity Enhancement by Water-Mediated Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
5U1H
 
 | | Crystal structure of the C-terminal peptidoglycan binding domain of OprF (PA1777) from Pseudomonas aeruginosa | | Descriptor: | (2R,6S)-2-amino-6-(carboxyamino)-7-{[(1R)-1-carboxyethyl]amino}-7-oxoheptanoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Watanabe, N, Stogios, P.J, Skarina, T, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-28 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the C-terminal peptidoglycan binding domain of OprF (PA1777) from Pseudomonas aeruginosa
To be published
|
|
7VIT
 
 | | Crystal structure of Au(400EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferritin light chain, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
8OK7
 
 | | Variant Surface Glycoprotein VSG558 NTD | | Descriptor: | Variant surface glycoprotein 558, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zeelen, J.P, Stebbins, C.E, van Straaten, M, Zhong, J. | | Deposit date: | 2023-03-27 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A structural classification of the variant surface glycoproteins of the African trypanosomey.
Plos Negl Trop Dis, 17, 2023
|
|
7NNI
 
 | |
5U1Q
 
 | | Grb7-SH2 with bicyclic peptide inhibitor | | Descriptor: | CHLORIDE ION, Growth factor receptor-bound protein 7, LYS-PHE-GLU-GLY-TYR-ASP-ASN-GLU-CST | | Authors: | Watson, G.M, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2016-11-28 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target.
J. Med. Chem., 60, 2017
|
|
5H3S
 
 | | apo form of GEMIN5-WD | | Descriptor: | GLYCEROL, Gem-associated protein 5 | | Authors: | Bharath, S.R, Tang, X, Song, H. | | Deposit date: | 2016-10-27 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for specific recognition of pre-snRNA by Gemin5
Cell Res., 26, 2016
|
|
8OMF
 
 | | Crystal structure of hKHK-C in complex with compound-4 | | Descriptor: | Ketohexokinase, SULFATE ION, compound | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structures of human and mouse ketohexokinase provide a structural basis for species- and isoform-selective inhibitor design.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7NLW
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2-(5-methoxy-1H-indol-3-yl)acetonitrile | | Descriptor: | 2-(5-methoxy-1~{H}-indol-3-yl)ethanenitrile, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
8S69
 
 | | Low pH (5.5) as-isolated MSOX movie series dataset 40 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [22.8 MGy] | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Horrell, S, Brondino, C.D, Eady, R.R, Jaho, S, Hough, M.A, Owen, A.L, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
5U9N
 
 | | Second Bromodomain of cdg4_1340 from Cryptosporidium parvum, complexed with bromosporine | | Descriptor: | Bromo domain containing protein, Bromosporine, SULFATE ION | | Authors: | Hou, C.F.D, Lin, Y.H, Loppnau, P, Hutchinson, A, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, Walker, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-12-16 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Second Bromodomain of cdg4_1340 from Cryptosporidium parvum, complexed with bromosporine
To be published
|
|
