2KCD
 
 | | Solution NMR structure of SSP0047 from Staphylococcus saprophyticus. Northeast Structural Genomics Consortium Target SyR6. | | Descriptor: | Uncharacterized protein SSP0047 | | Authors: | Ramelot, T.A, Ding, K, Chen, C.X, Jiang, M, Ciccosanti, C, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-19 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of SSP0047 from Staphylococcus saprophyticus. Northeast Structural Genomics Consortium Target SyR6.
To be Published
|
|
2KJR
 
 | | Solution NMR structure of the N-terminal Ubiquitin-like Domain from Tubulin-binding Cofactor B, CG11242, from Drosophila melanogaster. Northeast Structural Genomics Consortium Target FR629A (residues 8-92) | | Descriptor: | CG11242 | | Authors: | Ramelot, T.A, Cort, J.R, Shastry, R, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Swapna, G, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-08 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the N-terminal Ubiquitin-like Domain from
Tubulin-binding Cofactor B, CG11242, from Drosophila melanogaster. Northeast
Structural Genomics Consortium Target FR629A (residues 8-92)
To be Published
|
|
2KON
 
 | | NMR solution structure of CV_2116 from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CvT4(1-82) | | Descriptor: | Uncharacterized protein | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Garcia, M, Yee, A, Arrowsmith, C.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-24 | | Release date: | 2009-10-13 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of CV_2116 from Chromobacterium violaceum.Northeast Structural Genomics Consortium Target CvT4(1-82)
To be Published
|
|
2KVO
 
 | | Solution NMR structure of Photosystem II reaction center Psb28 protein from Synechocystis sp.(strain PCC 6803), Northeast Structural Genomics Consortium Target SgR171 | | Descriptor: | Photosystem II reaction center psb28 protein | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Wang, D, Ciccosanti, C, Hamilton, K, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-22 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of photosystem II reaction center protein Psb28 from Synechocystis sp. Strain PCC 6803.
Proteins, 79, 2011
|
|
2LF2
 
 | | Solution NMR structure of the AHSA1-like protein CHU_1110 from Cytophaga hutchinsonii, Northeast Structural Genomics Consortium Target ChR152 | | Descriptor: | Uncharacterized protein | | Authors: | Yang, Y, Ramelot, T.A, Lee, D, Ciccosanti, C, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the AHSA1-like protein CHU_1110 from Cytophaga hutchinsonii, Northeast Structural Genomics Consortium Target ChR152.
To be Published
|
|
2LCG
 
 | | Solution NMR structure of protein Rmet_5065 from Ralstonia metallidurans, Northeast Structural Genomics Consortium Target CrR115 | | Descriptor: | Uncharacterized protein | | Authors: | Ramelot, T.A, Yang, Y, Wang, D, Ciccosanti, C, Janjua, H, Nair, R, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein Rmet_5065 from Ralstonia metallidurans.
Northeast Structural Genomics Consortium Target CrR115.
To be Published
|
|
2L8V
 
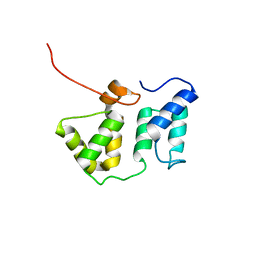 | | Solution NMR structure of the phycobilisome linker polypeptide domain of CpcC (20-153) from Thermosynechococcus elongatus, Northeast Structural Genomics Consortium Target TeR219A | | Descriptor: | Phycobilisome 32.1 kDa linker polypeptide, phycocyanin-associated, rod | | Authors: | Ramelot, T.A, Yang, Y, Cort, J.R, Lee, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-01-26 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the phycobilisome linker polypeptide domain of CpcC
(20-153) from Thermosynechococcus elongatus, Northeast Structural Genomics
Consortium Target TeR219A
To be Published
|
|
2LAK
 
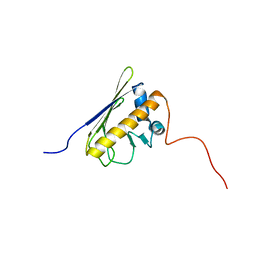 | | Solution NMR structure of the AHSA1-like protein RHE_CH02687 (1-152) from Rhizobium etli, Northeast Structural Genomics Consortium Target ReR242 | | Descriptor: | AHSA1-like protein RHE_CH02687 | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Wang, D, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-03-16 | | Release date: | 2011-04-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the AHSA1-like protein RHE_CH02687 (1-152) from Rhizobium etli, Northeast Structural Genomics Consortium Target ReR242
To be Published
|
|
2KZV
 
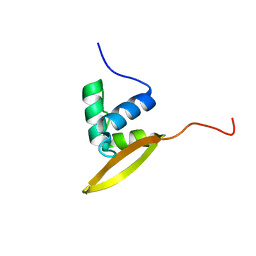 | | Solution NMR structure of CV_0373(175-257) protein from Chromobacterium violaceum, Northeast Structural Genomics Consortium Target CvR118A | | Descriptor: | Uncharacterized protein | | Authors: | Yang, Y, Ramelot, T.A, Wang, D, Ciccosanti, C, Mao, L, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-25 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of CV_0373(175-257) protein from Chromobacterium violaceum, Northeast Structural Genomics Consortium Target CvR118A.
To be Published
|
|
2LL8
 
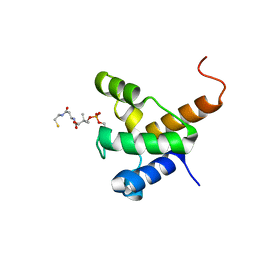 | | Solution NMR structure of the specialized holo-acyl carrier protein RPA2022 from Rhodopseudomonas palustris refined with NH RDCs, Northeast Structural Genomics Consortium Target RpR324 | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Specialized acyl carrier protein | | Authors: | Ramelot, T.A, Ni, S, Rossi, P, Yang, Y, Wang, H, Ciccosanti, C, Maglaqui, M, Janjua, H, Nair, R, Roset, B, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of a specialized acyl carrier protein essential for lipid A biosynthesis with very long-chain fatty acids in open and closed conformations.
Biochemistry, 51, 2012
|
|
2L09
 
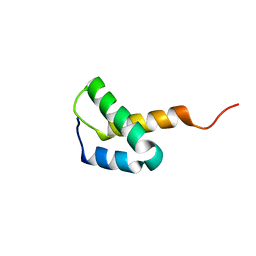 | | Solution NMR Structure of Protein asr4154 from Nostoc sp. PCC7120 Northeast Structural Genomics Consortium target ID NsR143 | | Descriptor: | Asr4154 protein | | Authors: | Feldmann, E.A, Yang, Y.A, Ramelot, T.A, Cort, J.R, Janjua, H, Ciccosanti, C.A, Lee, D, Acton, T.B, Xiao, R.A, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of asr4154 from Nostoc sp. PCC7120 Northeast Structural Genomics target ID Nsr143
To be Published
|
|
2JRS
 
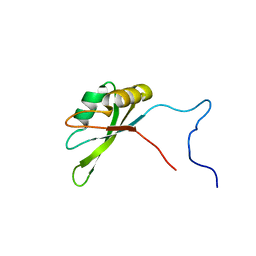 | | Solution NMR Structure of CAPER RRM2 Domain. Northeast Structural Genomics Target HR4730A | | Descriptor: | RNA-binding protein 39 | | Authors: | Rossi, P, Zhao, L, Nwosu, C, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-28 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of CAPER RRM2 Domain.
To be Published
|
|
2KOB
 
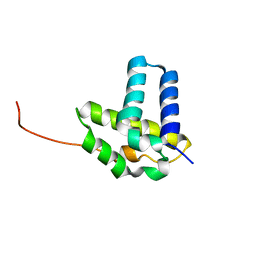 | | Solution NMR structure of CLOLEP_01837 (fragment 61-160) from Clostridium leptum. Northeast Structural Genomics Consortium Target QlR8A | | Descriptor: | Uncharacterized protein | | Authors: | Ramelot, T.A, Lee, D, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-15 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of CLOLEP_01837 (fragment 61-160) from Clostridium leptum. Northeast Structural Genomics Consortium Target QlR8A
To be Published
|
|
2KYI
 
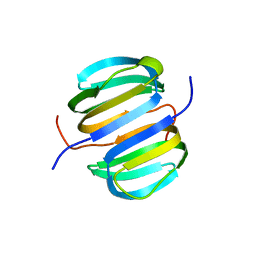 | | Solution NMR structure of Dsy0195(21-82) protein from Desulfitobacterium Hafniense. Northeast Structural Genomics Consortium Target DhR8C | | Descriptor: | Uncharacterized protein | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Wang, H, Ciccosanti, C, Foote, E.L, Jiang, M, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-05-27 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Combining NMR and EPR methods for homodimer protein structure determination.
J.Am.Chem.Soc., 132, 2010
|
|
2KRS
 
 | | Solution NMR structure of SH3 domain from CPF_0587 (fragment 415-479) from Clostridium perfringens. Northeast Structural Genomics Consortium (NESG) Target CpR74A. | | Descriptor: | Probable enterotoxin | | Authors: | Ramelot, T.A, Cort, J.R, Maglaqui, M, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-22 | | Release date: | 2010-01-26 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of SH3 domain from CPF_0587 (fragment 415-479) from Clostridium perfringens. Northeast Structural Genomics Consortium (NESG) Target CpR74A.
To be Published
|
|
2KIW
 
 | | Solution NMR structure of the domain N-terminal to the integrase domain of SH1003 from Staphylococcus haemolyticus. Northeast Structural Genomics Consortium Target ShR105F (64-166). | | Descriptor: | Int protein | | Authors: | Yang, Y, Ramelot, T.A, Belote, R.L, Foote, E.L, Janjua, H, Nair, R, Rost, B, Swapna, G, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-12 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the domain N-terminal to the integrase domain of SH1003 from Staphylococcus
haemolyticus. Northeast Structural Genomics Consortium Target ShR105F (64-166).
To be Published
|
|
2LGH
 
 | | Solution NMR structure of the AHSA1-like protein AHA_2358 from Aeromonas hydrophila refined with NH RDCs, Northeast Structural Genomics Consortium Target AhR99. | | Descriptor: | Uncharacterized protein | | Authors: | Ramelot, T.A, Yang, Y, Lee, H, Wang, D, Ciccosanti, C, Janjua, H, Nair, R, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-26 | | Release date: | 2011-08-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the AHSA1-like protein AHA_2358 from Aeromonas
hydrophila refined with NH RDCs. Northeast Structural Genomics Consortium Target AhR99.
To be Published
|
|
2KT8
 
 | | Solution NMR structure of the CPE1231(468-535) protein from Clostridium perfringens, Northeast Structural Genomics Consortium Target CpR82B | | Descriptor: | Probable surface protein | | Authors: | Yang, Y, Ramelot, T.A, Lee, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-21 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the CPE1231(468-535) protein from Clostridium perfringens, Northeast Structural Genomics Consortium Target CpR82B
To be Published
|
|
2K9Q
 
 | | Solution NMR structure of HTH_XRE family transcriptional regulator BT_p548217 from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR244. | | Descriptor: | uncharacterized protein | | Authors: | Ramelot, T.A, Cort, J.R, Zhao, L, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-23 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of HTH_XRE family transcriptional regulator BT_p548217 from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR244.
To be Published
|
|
2KHD
 
 | | Solution NMR structure of VC_A0919 from Vibrio cholerae. Northeast Structural Genomics Consortium Target VcR52 | | Descriptor: | Uncharacterized protein VC_A0919 | | Authors: | Ramelot, T.A, Cort, J.R, Wang, H, Ciccosanti, C, Jiang, M, Liu, J, Rost, B, Swapna, G.V.T, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-04-02 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of VC_A0919 from Vibrio cholerae. Northeast Structural Genomics Consortium Target VcR52.
To be Published
|
|
2KPI
 
 | | Solution NMR structure of Streptomyces coelicolor SCO3027 modeled with Zn+2 bound, Northeast Structural Genomics Consortium Target RR58 | | Descriptor: | Uncharacterized protein SCO3027, ZINC ION | | Authors: | Ramelot, T.A, Cort, J.R, Garcia, M, Yee, A, Arrowmith, C.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Streptomyces coelicolor SCO3027 modeled with Zn+2 bound, Northeast Structural Genomics Consortium Target RR58
To be Published
|
|
2KKL
 
 | | Solution NMR structure of FHA domain of Mb1858 from Mycobacterium bovis. Northeast Structural Genomics Consortium Target MbR243C (24-155). | | Descriptor: | Uncharacterized protein Mb1858 | | Authors: | Yang, Y, Ramelot, T.A, Wang, D, Foote, E.L, Jiang, M, Nair, R, Rost, B, Swapna, G, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-25 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of FHA domain of Mb1858 from Mycobacterium bovis. Northeast Structural Genomics Consortium Target MbR243C (24-155).
To be Published
|
|
2KS0
 
 | | Solution NMR structure of the Q251Q8_DESHY(21-82) protein from Desulfitobacterium Hafniense, Northeast Structural Genomics Consortium Target DhR8C | | Descriptor: | Uncharacterized protein | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Wang, H, Ciccosanti, C, Foote, E.L, Jiang, M, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-23 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Combining NMR and EPR methods for homodimer protein structure determination.
J.Am.Chem.Soc., 132, 2010
|
|
2LK2
 
 | | Solution NMR structure of homeobox domain (171-248) of human homeobox protein TGIF1, Northeast Structural Genomics Consortium Target HR4411B | | Descriptor: | Homeobox protein TGIF1 | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Shastry, R, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-30 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of homeobox domain (171-248) of human homeobox protein TGIF1, Northeast Structural Genomics Consortium Target HR4411B.
To be Published
|
|
2LPK
 
 | | Solution NMR of the specialized apo-acyl carrier protein (RPA2022) from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR324 | | Descriptor: | Specialized acyl carrier protein | | Authors: | Ramelot, T.A, Rossi, P, Yang, Y, Lee, H, Ertekin, A, Wang, H, Ciccosanti, C, Kohan, E, Maglaqui, M, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-14 | | Release date: | 2012-02-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a specialized acyl carrier protein essential for lipid A biosynthesis with very long-chain fatty acids in open and closed conformations.
Biochemistry, 51, 2012
|
|
