4CN3
 
 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to the Human Gde1SpA Response Element | | Descriptor: | 5'-D(*CP*TP*AP*GP*TP*TP*CP*AP*AP*AP*GP*TP*TP*CP *AP*CP*A)-3', 5'-D(*TP*GP*TP*GP*AP*AP*CP*TP*TP*TP*GP*AP*AP*CP *TP*AP*G)-3', RETINOIC ACID RECEPTOR RXR-ALPHA, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2014-01-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Natural Promoter Recognition by the Retinoid X Nuclear Receptor.
Sci.Rep., 5, 2015
|
|
1UOL
 
 | |
1R4I
 
 | | Crystal Structure of Androgen Receptor DNA-Binding Domain Bound to a Direct Repeat Response Element | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*TP*CP*AP*AP*GP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*TP*CP*TP*TP*GP*AP*TP*GP*TP*TP*CP*TP*GP*G)-3', Androgen receptor, ... | | Authors: | Shaffer, P.L, Jivan, A, Dollins, D.E, Claessens, F, Gewirth, D.T. | | Deposit date: | 2003-10-06 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of androgen receptor binding to selective androgen response elements.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1AJY
 
 | | STRUCTURE AND MOBILITY OF THE PUT3 DIMER: A DNA PINCER, NMR, 13 STRUCTURES | | Descriptor: | PUT3, ZINC ION | | Authors: | Walters, K.J, Dayie, K.T, Reece, R.J, Ptashne, M, Wagner, G. | | Deposit date: | 1997-05-12 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and mobility of the PUT3 dimer.
Nat.Struct.Biol., 4, 1997
|
|
1FBQ
 
 | |
3MGR
 
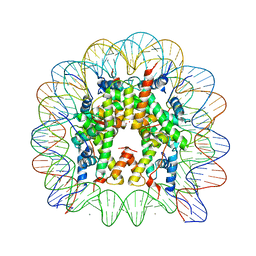 | | Binding of Rubidium ions to the Nucleosome Core Particle | | Descriptor: | CHLORIDE ION, DNA (147-MER), Histone H2A, ... | | Authors: | Mohideen, K, Muhammad, R, Davey, C.A. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Perturbations in nucleosome structure from heavy metal association.
Nucleic Acids Res., 38, 2010
|
|
3MGP
 
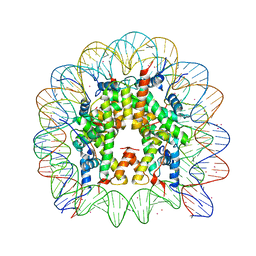 | | Binding of Cobalt ions to the Nucleosome Core Particle | | Descriptor: | CHLORIDE ION, COBALT (II) ION, DNA (147-MER), ... | | Authors: | Mohideen, K, Muhammad, R, Davey, C.A. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Perturbations in nucleosome structure from heavy metal association.
Nucleic Acids Res., 38, 2010
|
|
1FBU
 
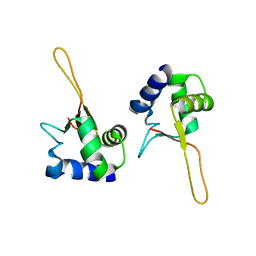 | | HEAT SHOCK TRANSCRIPTION FACTOR DNA BINDING DOMAIN | | Descriptor: | HEAT SHOCK FACTOR PROTEIN | | Authors: | Hardy, J.A, Nelson, H.C.M. | | Deposit date: | 2000-07-16 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Proline in alpha-helical kink is required for folding kinetics but not for kinked structure, function, or stability of heat shock transcription factor.
Protein Sci., 9, 2000
|
|
4ZM2
 
 | |
7NVX
 
 | | TFIIH in a post-translocated state (with ADP-BeF3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CDK-activating kinase assembly factor MAT1, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
7NVW
 
 | | TFIIH in a pre-translocated state (without ADP-BeF3) | | Descriptor: | CDK-activating kinase assembly factor MAT1, General transcription and DNA repair factor IIH helicase subunit XPB, General transcription factor IIE subunit 1, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
5N9F
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with ssDNA CpG_A | | Descriptor: | CG9323, isoform A, CHLORIDE ION, ... | | Authors: | Chen, W.-F, Rety, S, Hai-Lei Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.969 Å) | | Cite: | Structural and mechanistic insights into DHX36-mediated innate immunity and G-quadruplex unfolding
To Be Published
|
|
6QVW
 
 | | Solution structure of the free FOXO1 DNA binding domain | | Descriptor: | Forkhead box protein O1 | | Authors: | Psenakova, K, Obsil, T, Veverka, V, Obsilova, V, Kohoutova, K. | | Deposit date: | 2019-03-05 | | Release date: | 2019-09-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Forkhead Domains of FOXO Transcription Factors Differ in both Overall Conformation and Dynamics.
Cells, 8, 2019
|
|
3MGS
 
 | |
1GDT
 
 | |
3MGQ
 
 | | Binding of Nickel ions to the Nucleosome Core Particle | | Descriptor: | CHLORIDE ION, DNA (147-MER), Histone H2A, ... | | Authors: | Mohideen, K, Muhammad, R, Davey, C.A. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Perturbations in nucleosome structure from heavy metal association.
Nucleic Acids Res., 38, 2010
|
|
1H38
 
 | | Structure of a T7 RNA polymerase elongation complex at 2.9A resolution | | Descriptor: | 5'-D(*GP*GP*GP*AP*AP*TP*CP*GP*AP*CP *AP*TP*CP*GP*CP*CP*GP*C)-3', 5'-D(*GP*TP*CP*GP*AP*TP*TP*CP*CP*CP)-3', 5'-R(*AP*AP*CP*UP*GP*CP*GP*GP*CP*GP *AP*U)-3', ... | | Authors: | Tahirov, T.H, Temyakov, D, Anikin, M, Patlan, V, McAllister, W.T, Vassylyev, D.G, Yokoyama, S. | | Deposit date: | 2002-08-24 | | Release date: | 2002-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a T7 RNA Polymerase Elongation Complex at 2.9 A Resolution
Nature, 420, 2002
|
|
2B0L
 
 | | C-terminal DNA binding domain of transcriptional pleiotropic repressor CodY. | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor codY | | Authors: | Levdikov, V.M, Blagova, E, Joseph, P, Sonenshein, A.L, Wilkinson, A.J. | | Deposit date: | 2005-09-14 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of CodY, a GTP- and Isoleucine-responsive Regulator of Stationary Phase and Virulence in Gram-positive Bacteria.
J.Biol.Chem., 281, 2006
|
|
8THO
 
 | |
4R4I
 
 | | Structure of RPA70N in complex with 5-(4-((6-(5-carboxyfuran-2-yl)-1-thioxo-3,4-dihydroisoquinolin-2(1H)-yl)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-(4-{[6-(5-carboxyfuran-2-yl)-1-thioxo-3,4-dihydroisoquinolin-2(1H)-yl]methyl}phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
4R4O
 
 | | Crystal structure of RPA70N in complex with 5-(4-((4-(5-carboxyfuran-2-yl)benzyl)carbamothioyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-(4-{[4-(5-carboxyfuran-2-yl)benzyl]carbamothioyl}phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
4R4T
 
 | | Crystal Structure of RPA70N in complex with 5-(4-((4-(5-carboxyfuran-2-yl)phenylthioamido)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-{4-[({[4-(5-carboxyfuran-2-yl)phenyl]carbonothioyl}amino)methyl]phenyl}-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
4R4Q
 
 | | Crystal structure of RPA70N in complex with 5-(3-((N-(4-(5-carboxyfuran-2-yl)benzyl)acetamido)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-[4-({acetyl[4-(5-carboxyfuran-2-yl)benzyl]amino}methyl)phenyl]-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
8E67
 
 | |
8E66
 
 | | ETV6 H396Y variant bound to DNA containing the sequence GGAA | | Descriptor: | CACODYLATE ION, Complementary 15 bp strand, GGAA-containing 15 bp DNA, ... | | Authors: | Scheu, K, Chan, A.C, Murphy, M.E, McIntosh, L.P. | | Deposit date: | 2022-08-22 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The functional role of histidine within the ETV6 ETS domain
to be published
|
|
