9JO5
 
 | | Structure of isw1-nucleosome complex in ADP-B state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JNT
 
 | | Structure of isw1-nucleosome complex in ADP* state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
6A5U
 
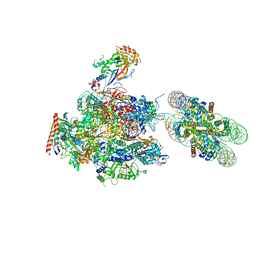 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA, tilt conformation | | Descriptor: | DNA (198-MER), DNA (40-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
9JO2
 
 | | Structure of isw1-nucleosome complex in Apo* state | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JNX
 
 | | Structure of isw1-nucleosome complex in ADP*+ state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JNZ
 
 | | Structure of isw1-nucleosome complex in Apo state | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JNW
 
 | | Structure of isw1-nucleosome complex in ADP+ state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (147-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JNU
 
 | | Structure of isw1-nucleosome complex in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JAO
 
 | | The structure of SMARCAD1 bound to the hexasome in the presence of ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (157-MER), ... | | Authors: | Chen, Z.C, Hu, P.J, Sia, Y.Y, Chen, K.J, Xia, X. | | Deposit date: | 2024-08-25 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Subnucleosome preference of human chromatin remodeller SMARCAD1.
Nature, 2025
|
|
9K3Z
 
 | |
9K40
 
 | |
9K42
 
 | |
9K41
 
 | |
9K43
 
 | |
7KBF
 
 | |
7KBE
 
 | |
7KBD
 
 | |
2FUI
 
 | |
8F8Y
 
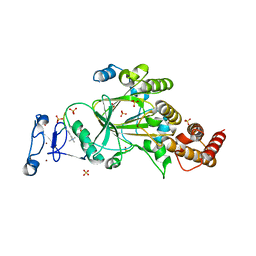 | | PHF2 (PHD+JMJ) in Complex with VRK1 N-Terminal Peptide | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase PHF2, SULFATE ION, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2022-11-22 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A complete methyl-lysine binding aromatic cage constructed by two domains of PHF2.
J.Biol.Chem., 299, 2022
|
|
1S32
 
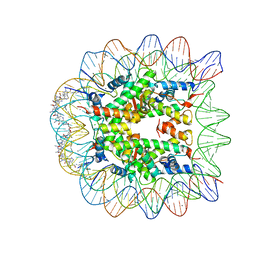 | | Molecular Recognition of the Nucleosomal 'Supergroove' | | Descriptor: | 2-(2-CARBAMOYLMETHOXY-ETHOXY)-ACETAMIDE, 3-AMINO-(DIMETHYLPROPYLAMINE), 4-AMINO-(1-METHYLIMIDAZOLE)-2-CARBOXYLIC ACID, ... | | Authors: | Edayathumangalam, R.S, Weyermann, P, Gottesfeld, J.M, Dervan, P.B, Luger, K. | | Deposit date: | 2004-01-12 | | Release date: | 2004-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular Recognition of the Nucleosomal 'Supergroove'
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3Q66
 
 | |
1M18
 
 | | LIGAND BINDING ALTERS THE STRUCTURE AND DYNAMICS OF NUCLEOSOMAL DNA | | Descriptor: | Histone H2A.1, Histone H2B.1, Histone H3.2, ... | | Authors: | Suto, R.K, Edayathumangalam, R.S, White, C.L, Melander, C, Gottesfeld, J.M, Dervan, P.B, Luger, K. | | Deposit date: | 2002-06-18 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structures of Nucleosome Core Particles in Complex with Minor Groove DNA-binding Ligands
J.Mol.Biol., 326, 2003
|
|
2F6N
 
 | |
2KWJ
 
 | | Solution structures of the double PHD fingers of human transcriptional protein DPF3 bound to a histone peptide containing acetylation at lysine 14 | | Descriptor: | Histone peptide, ZINC ION, Zinc finger protein DPF3 | | Authors: | Zeng, L, Zhang, Q, Li, S, Plotnikov, A.N, Walsh, M.J, Zhou, M. | | Deposit date: | 2010-04-12 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b.
Nature, 466, 2010
|
|
3Q68
 
 | |
