4HTB
 
 | | tRNA-guanine transglycosylase Y330C mutant in space group C2 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, ZINC ION, ... | | Authors: | Jakobi, S, Klebe, G. | | Deposit date: | 2012-11-01 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | What Glues a Homodimer Together: Systematic Analysis of the Stabilizing Effect of an Aromatic Hot Spot in the Protein-Protein Interface of the tRNA-Modifying Enzyme Tgt.
Acs Chem.Biol., 10, 2015
|
|
2YB1
 
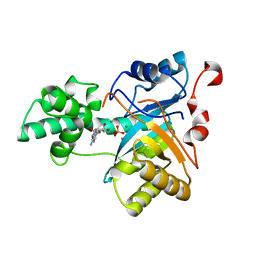 | | Structure of an amidohydrolase from Chromobacterium violaceum (EFI target EFI-500202) with bound Mn, AMP and phosphate. | | Descriptor: | ADENOSINE MONOPHOSPHATE, AMIDOHYDROLASE, MANGANESE (II) ION, ... | | Authors: | Vetting, M.W, Hillerich, B, Foti, R, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-02-25 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Prospecting for Unannotated Enzymes: Discovery of a 3',5'-Nucleotide Bisphosphate Phosphatase within the Amidohydrolase Superfamily.
Biochemistry, 53, 2014
|
|
6BNO
 
 | | Structure of bare actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gurel, P.S, Alushin, G.A. | | Deposit date: | 2017-11-17 | | Release date: | 2018-01-10 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Cryo-EM structures reveal specialization at the myosin VI-actin interface and a mechanism of force sensitivity.
Elife, 6, 2017
|
|
6BNQ
 
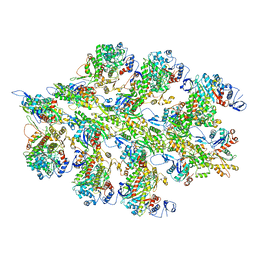 | |
3NF2
 
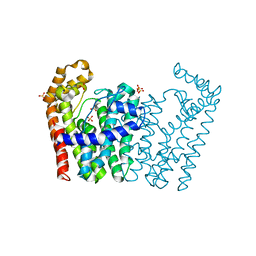 | | Crystal structure of polyprenyl synthetase from Streptomyces coelicolor A3(2) | | Descriptor: | Putative polyprenyl synthetase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3MZV
 
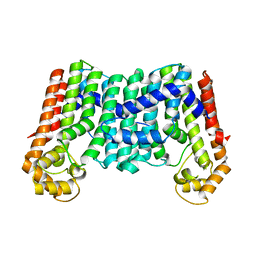 | | Crystal structure of a decaprenyl diphosphate synthase from Rhodobacter capsulatus | | Descriptor: | Decaprenyl diphosphate synthase | | Authors: | Quartararo, C.E, Patskovsky, Y, Bonanno, J.B, Rutter, M, Bain, K.T, Chang, S, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-13 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5VW6
 
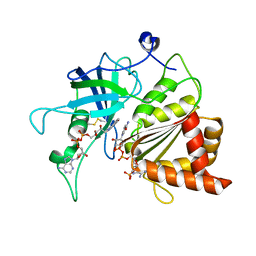 | | NADP+ soak of Y316A mutant of corn root ferredoxin:NADP+ reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, MAGNESIUM ION, ... | | Authors: | Kean, K.M, Carpenter, R.A, Hall, A.R, Karplus, P.A. | | Deposit date: | 2017-05-21 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution studies of hydride transfer in the ferredoxin:NADP(+) reductase superfamily.
FEBS J., 284, 2017
|
|
6BNP
 
 | |
6BNW
 
 | |
3N5G
 
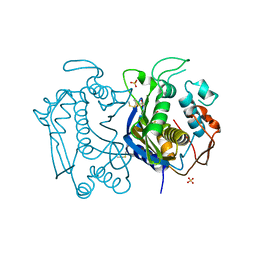 | | Crystal Structure of histidine-tagged human thymidylate synthase | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Pozzi, C, Cardinale, D, Guaitoli, G, Tondi, D, Luciani, R, Myllykallio, H, Ferrari, S, Costi, M.P, Mangani, S. | | Deposit date: | 2010-05-25 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Protein-protein interface-binding peptides inhibit the cancer therapy target human thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4I37
 
 | |
2PYY
 
 | | Crystal Structure of the GluR0 ligand-binding core from Nostoc punctiforme in complex with (L)-glutamate | | Descriptor: | GLUTAMIC ACID, Ionotropic glutamate receptor bacterial homologue | | Authors: | Lee, J.H, Kang, G.B, Lim, H.-H, Ree, M, Park, C.-S, Eom, S.H. | | Deposit date: | 2007-05-17 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the GluR0 ligand-binding core from Nostoc punctiforme in complex with L-glutamate: structural dissection of the ligand interaction and subunit interface.
J.Mol.Biol., 376, 2008
|
|
3EA2
 
 | | Crystal Structure of the Myo-inositol bound Y247S/Y251S Mutant of Phosphatidylinositol-Specific Phospholipase C from Bacillus Thuringiensis | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, ZINC ION | | Authors: | Shi, X, Shao, C, Zhang, X, Zambonelli, C, Redfied, A.G, Head, J.F, Seaton, B.A, Roberts, M.F. | | Deposit date: | 2008-08-24 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Modulation of bacillus thuringiensis phosphatidylinositol-specific phospholipase C activity by mutations in the putative dimerization interface.
J.Biol.Chem., 284, 2009
|
|
3SQ5
 
 | |
1VAR
 
 | |
4I6I
 
 | |
6BNV
 
 | |
4IL8
 
 | | Crystal structure of an H329A mutant of p. aeruginosa PMM/PGM | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphomannomutase/phosphoglucomutase | | Authors: | Lee, Y, Mehra-Chaudhary, R, Furdui, C, Beamer, L. | | Deposit date: | 2012-12-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of an essential active-site residue in the alpha-D-phosphohexomutase enzyme superfamily.
Febs J., 280, 2013
|
|
3SQ3
 
 | |
5VW7
 
 | | NADPH soak of Y316A mutant of corn root ferredoxin:NADP+ reductase | | Descriptor: | ACETATE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Kean, K.M, Carpenter, R.A, Hall, A.R, Karplus, P.A. | | Deposit date: | 2017-05-21 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.608 Å) | | Cite: | High-resolution studies of hydride transfer in the ferredoxin:NADP(+) reductase superfamily.
FEBS J., 284, 2017
|
|
5VW3
 
 | | NADP+ soak of Y316S mutant of corn root ferredoxin:NADP+ reductase | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Kean, K.M, Carpenter, R.A, Hall, A.R, Karplus, P.A. | | Deposit date: | 2017-05-21 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | High-resolution studies of hydride transfer in the ferredoxin:NADP(+) reductase superfamily.
FEBS J., 284, 2017
|
|
6BNU
 
 | |
5VW2
 
 | | NADPH soak of Y316S mutant of corn root ferredoxin:NADP+ reductase | | Descriptor: | ACETATE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Kean, K.M, Carpenter, R.A, Hall, A.R, Karplus, P.A. | | Deposit date: | 2017-05-21 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | High-resolution studies of hydride transfer in the ferredoxin:NADP(+) reductase superfamily.
FEBS J., 284, 2017
|
|
5VWB
 
 | | Y316F mutant of corn root ferredoxin:NADP+ reductase in alternate space group | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, MAGNESIUM ION, ... | | Authors: | Kean, K.M, Carpenter, R.A, Hall, A.R, Karplus, P.A. | | Deposit date: | 2017-05-21 | | Release date: | 2017-08-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution studies of hydride transfer in the ferredoxin:NADP(+) reductase superfamily.
FEBS J., 284, 2017
|
|
5VW5
 
 | | Nicotinamide soak of Y316A mutant of corn root ferredoxin:NADP+ reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, MAGNESIUM ION, ... | | Authors: | Kean, K.M, Carpenter, R.A, Hall, A.R, Karplus, P.A. | | Deposit date: | 2017-05-21 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | High-resolution studies of hydride transfer in the ferredoxin:NADP(+) reductase superfamily.
FEBS J., 284, 2017
|
|
