4YHR
 
 | | Crystal Structure of Yeast Proliferating Cell Nuclear Antigen | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Litman, J.M, Nguyen, V.Q, Kondratick, C.M, Powers, K.T, Schnieders, M.J, Washington, M.T. | | Deposit date: | 2015-02-27 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9502 Å) | | Cite: | Dead-End Elimination with a Polarizable Force Field Repacks PCNA Models from Low-Resolution X-ray Diffraction into Atomic Resolution Structures
To be published
|
|
2CII
 
 | | The crystal structure of H-2Db complexed with a partial peptide epitope suggests an MHC Class I assembly-intermediate | | Descriptor: | BETA-2-MICROGLOBULIN, GLYCEROL, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN D-B ALPHA CHAIN, ... | | Authors: | Glithero, A, Tormo, J, Doering, K, Kojima, M, Jones, E.Y, Elliott, T. | | Deposit date: | 2006-03-21 | | Release date: | 2006-03-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of H-2D(b) complexed with a partial peptide epitope suggests a major histocompatibility complex class I assembly intermediate.
J. Biol. Chem., 281, 2006
|
|
7C5Z
 
 | |
4CEO
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (2S)-2-(3-hydroxy-3-oxopropyl)-6-[[[2-[(4-methoxyphenyl)methylcarbamoyl]phenyl]methyl-methyl-amino]methyl]-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-11 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
4CEF
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (2S)-2-(3-hydroxy-3-oxopropyl)-6-[[methyl-[[2-[(phenylmethyl)carbamoyl]phenyl]methyl]amino]methyl]-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-11 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
4CEE
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (2S)-6-[[[2-[[(2S)-butan-2-yl]carbamoyl]phenyl]methyl-methyl-amino]methyl]-2-(3-hydroxy-3-oxopropyl)-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-11 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
6G5R
 
 | |
1ZY2
 
 | | Crystal structure of the phosphorylated receiver domain of the transcription regulator NtrC1 from Aquifex aeolicus | | Descriptor: | MAGNESIUM ION, transcriptional regulator NtrC1 | | Authors: | Doucleff, M, Chen, B, Maris, A.E, Wemmer, D.E, Kondrashkina, E, Nixon, B.T. | | Deposit date: | 2005-06-09 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Negative regulation of AAA + ATPase assembly by two component receiver domains: a transcription activation mechanism that is conserved in mesophilic and extremely hyperthermophilic bacteria
J.Mol.Biol., 353, 2005
|
|
8PRK
 
 | | THE R78K AND D117E ACTIVE SITE VARIANTS OF SACCHAROMYCES CEREVISIAE SOLUBLE INORGANIC PYROPHOSPHATASE: STRUCTURAL STUDIES AND MECHANISTIC IMPLICATIONS | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, PROTEIN (INORGANIC PYROPHOSPHATASE) | | Authors: | Tuominen, V, Heikinheimo, P, Kajander, T, Torkkel, T, Hyytia, T, Kapyla, J, Lahti, R, Cooperman, B.S, Goldman, A. | | Deposit date: | 1998-09-16 | | Release date: | 1998-12-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The R78K and D117E active-site variants of Saccharomyces cerevisiae soluble inorganic pyrophosphatase: structural studies and mechanistic implications.
J.Mol.Biol., 284, 1998
|
|
5T9D
 
 | |
5A67
 
 | |
5A5Y
 
 | | Crystal structure of AtTTM3 in complex with tripolyphosphate and magnesium ion (form A) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, TRIPHOSPHATE, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
2VE6
 
 | | Crystal structure of a Murine MHC class I H2-Db molecule in complex with a photocleavable peptide | | Descriptor: | BETA-2-MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN D-B ALPHA CHAIN, SENDAI VIRUS EPITOPE RESIDUES 324-332 MODIFIED AT P7 | | Authors: | Grotenbreg, G.M, Roan, N.R, Guillen, E, Meijers, R, Wang, J.H, Bell, G.W, Starnbach, M.N, Ploegh, H.L. | | Deposit date: | 2007-10-17 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of Cd8+ T Cell Epitopes in Chlamydia Trachomatis Infection Through Use of Caged Class I Mhc Tetramers.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6GT7
 
 | |
5VHD
 
 | | DHX36 with an N-terminal truncation bound to ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DEAH (Asp-Glu-Ala-His) box polypeptide 36, TETRAFLUOROALUMINATE ION | | Authors: | Chen, M, Ferre-D'Amare, A. | | Deposit date: | 2017-04-13 | | Release date: | 2018-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36.
Nature, 558, 2018
|
|
5VHC
 
 | | DHX36 with an N-terminal truncation bound to ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DEAH (Asp-Glu-Ala-His) box polypeptide 36, ... | | Authors: | Chen, M, Ferre-D'Amare, A. | | Deposit date: | 2017-04-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36.
Nature, 558, 2018
|
|
5VHA
 
 | | DHX36 with an N-terminal truncation | | Descriptor: | DEAH (Asp-Glu-Ala-His) box polypeptide 36 | | Authors: | Chen, M, Ferre-D'Amare, A. | | Deposit date: | 2017-04-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36.
Nature, 558, 2018
|
|
5ZUT
 
 | | Crystal Structure of Yeast PCNA in Complex with N24 Peptide | | Descriptor: | N24, Proliferating cell nuclear antigen | | Authors: | Cheng, X.Y, Kuang, X.L, Zhou, Y, Xia, X.M, SU, Z.D. | | Deposit date: | 2018-05-08 | | Release date: | 2018-05-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal Structure of Yeast PCNA in Complex with N24 Peptide
To Be Published
|
|
1A1O
 
 | | MHC CLASS I MOLECULE B*5301 COMPLEXED WITH PEPTIDE LS6 (KPIVQYDNF) FROM THE MALARIA PARASITE P. FALCIPARUM | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, BW-53 B*5301 alpha chain, ... | | Authors: | Smith, K.J, Reid, S.W, Harlos, K, Mcmichael, A.J, Stuart, D.I, Bell, J.I, Jones, E.Y. | | Deposit date: | 1997-12-11 | | Release date: | 1998-04-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bound water structure and polymorphic amino acids act together to allow the binding of different peptides to MHC class I HLA-B53.
Immunity, 4, 1996
|
|
6W9W
 
 | | R80A PCNA mutant defective in BIR | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Ripley, B.M, Washington, M.T. | | Deposit date: | 2020-03-23 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural analysis of PCNA mutant proteins defective in BIR, FF248-249AA and R80A
To be Published
|
|
6WAC
 
 | |
1A1M
 
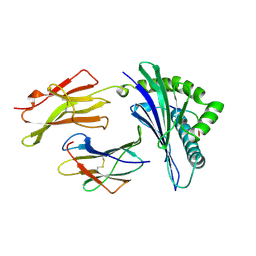 | | MHC CLASS I MOLECULE B*5301 COMPLEXED WITH PEPTIDE TPYDINQML FROM GAG PROTEIN OF HIV2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, BW-53 B*5301 alpha chain, ... | | Authors: | Smith, K.J, Reid, S.W, Harlos, K, Mcmichael, A.J, Stuart, D.I, Bell, J.I, Jones, E.Y. | | Deposit date: | 1997-12-11 | | Release date: | 1998-04-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bound water structure and polymorphic amino acids act together to allow the binding of different peptides to MHC class I HLA-B53.
Immunity, 4, 1996
|
|
1A1N
 
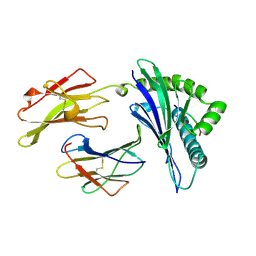 | | MHC CLASS I MOLECULE B*3501 COMPLEXED WITH PEPTIDE VPLRPMTY FROM THE NEF PROTEIN (75-82) OF HIV1 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, BW-53 B*5301 alpha chain, ... | | Authors: | Smith, K.J, Reid, S.W, Stuart, D.I, Mcmichael, A.J, Jones, E.Y, Bell, J.I. | | Deposit date: | 1997-12-11 | | Release date: | 1998-04-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An altered position of the alpha 2 helix of MHC class I is revealed by the crystal structure of HLA-B*3501.
Immunity, 4, 1996
|
|
6GVU
 
 | |
5A66
 
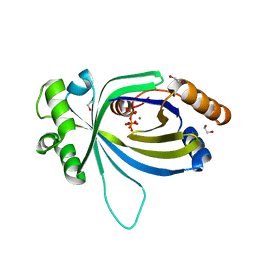 | | Crystal structure of AtTTM3 in complex with tripolyphosphate and manganese ion (form A) | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, TRIPHOSPHATE, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
