2HUA
 
 | |
2HVA
 
 | | Solution Structure of the haem-binding protein p22HBP | | Descriptor: | Heme-binding protein 1 | | Authors: | Gell, D.A, Mackay, J.P, Westman, B.J, Liew, C.K, Gorman, D. | | Deposit date: | 2006-07-28 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Novel Haem-binding Interface in the 22 kDa Haem-binding Protein p22HBP.
J.Mol.Biol., 362, 2006
|
|
2LD1
 
 | |
2BUS
 
 | |
2BEG
 
 | | 3D Structure of Alzheimer's Abeta(1-42) fibrils | | Descriptor: | Amyloid beta A4 protein | | Authors: | Luhrs, T, Ritter, C, Adrian, M, Riek-Loher, D, Bohrmann, B, Dobeli, H, Schubert, D, Riek, R. | | Deposit date: | 2005-10-24 | | Release date: | 2005-11-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 3D structure of Alzheimer's amyloid-{beta}(1-42) fibrils.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2C55
 
 | | Solution Structure of the Human Immunodeficiency Virus Type 1 p6 Protein | | Descriptor: | PROTEIN P6 | | Authors: | Fossen, T, Wray, V, Bruns, K, Rachmat, J, Henklein, P, Tessmer, U, Maczurek, A, Klinger, P, Schubert, U. | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Human Immunodeficiency Virus Type 1 P6 Protein.
J.Biol.Chem., 280, 2005
|
|
2BN8
 
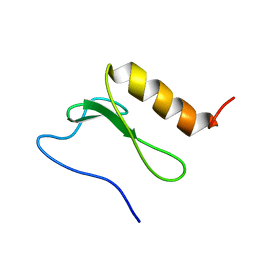 | | Solution Structure and interactions of the E .coli Cell Division Activator Protein CedA | | Descriptor: | CELL DIVISION ACTIVATOR CEDA | | Authors: | Chen, H.A, Simpson, P, Huyton, T, Roper, D, Matthews, S. | | Deposit date: | 2005-03-22 | | Release date: | 2006-12-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Interactions of the Escherichia Coli Cell Division Activator Protein Ceda.
Biochemistry, 44, 2005
|
|
2CMM
 
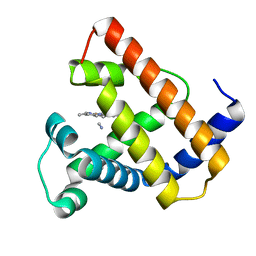 | | STRUCTURAL ANALYSIS OF THE MYOGLOBIN RECONSTITUTED WITH IRON PORPHINE | | Descriptor: | CYANIDE ION, MYOGLOBIN, PORPHYRIN FE(III) | | Authors: | Sato, T, Tanaka, N, Moriyama, H, Igarashi, N, Neya, S, Funasaki, N, Iizuka, T, Shiro, Y. | | Deposit date: | 1993-12-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the myoglobin reconstituted with iron porphine.
J.Biol.Chem., 268, 1993
|
|
2BBI
 
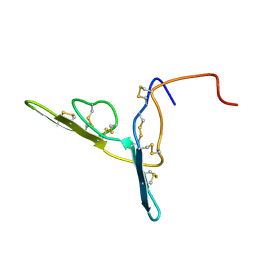 | |
2F52
 
 | |
2CRD
 
 | | ANALYSIS OF SIDE-CHAIN ORGANIZATION ON A REFINED MODEL OF CHARYBDOTOXIN: STRUCTURAL AND FUNCTIONAL IMPLICATIONS | | Descriptor: | CHARYBDOTOXIN | | Authors: | Bontems, F, Roumestand, C, Gilquin, B, Menez, A, Toma, F. | | Deposit date: | 1993-02-17 | | Release date: | 1993-07-15 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Analysis of side-chain organization on a refined model of charybdotoxin: structural and functional implications.
Biochemistry, 31, 1992
|
|
2G3S
 
 | | RNA structure containing GU base pairs | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*GP*CP*C)-3', MAGNESIUM ION | | Authors: | Jang, S.B, Hung, L.W, Jeong, M.S, Holbrook, E.L, Chen, X, Turner, D.H, Holbrook, S.R. | | Deposit date: | 2006-02-20 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | The crystal structure at 1.5 angstroms resolution of an RNA octamer duplex containing tandem G.U basepairs
Biophys.J., 90, 2006
|
|
2GA5
 
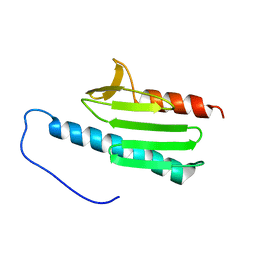 | | yeast frataxin | | Descriptor: | Frataxin homolog, mitochondrial | | Authors: | He, Y, Alam, S.L, Proteasa, S.V, Zhang, Y, Lesuisse, E, Dancis, A. | | Deposit date: | 2006-03-07 | | Release date: | 2006-03-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Yeast Frataxin Solution Structure, Iron Binding and Ferrochelatase Interaction
Biochemistry, 43, 2004
|
|
6KHU
 
 | |
2JU7
 
 | | Solution-State Structures of Oleate-Liganded LFABP, Protein Only | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | He, Y, Yang, X, Wang, H, Estephan, R, Francis, F, Kodukula, S, Storch, J, Stark, R.E. | | Deposit date: | 2007-08-15 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution-State Molecular Structure of Apo and Oleate-Liganded Liver Fatty Acid-Binding Protein
Biochemistry, 46, 2007
|
|
2JUH
 
 | |
2KFD
 
 | | Prp40 FF4 domain | | Descriptor: | Pre-mRNA-processing protein PRP40 | | Authors: | Bonet, R, Ruiz, L, Morales, B, Macias, M. | | Deposit date: | 2009-02-18 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fourth FF domain of yeast Prp40 splicing factor.
Proteins, 77, 2009
|
|
2KLZ
 
 | |
2K5E
 
 | | SOLUTION STRUCTURE OF PUTATIVE UNCHARACTERIZED PROTEIN GSU1278 FROM METHANOCALDOCOCCUS JANNASCHII, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM (NESG) TARGET GsR195 | | Descriptor: | uncharacterized protein | | Authors: | Liu, G, Zhao, L, Ciccosanti, C, Jiang, M, Xiao, R, Swapna, G, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-27 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE OF PUTATIVE UNCHARACTERIZED PROTEIN GSU1278 FROM METHANOCALDOCOCCUS JANNASCHII, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM (NESG) TARGET GsR195
To be Published
|
|
2KES
 
 | |
4NMQ
 
 | |
4NMV
 
 | |
2KBG
 
 | | Solution structure of the second Fibronectin type-III module of NCAM2 | | Descriptor: | Neural cell adhesion molecule 2 | | Authors: | Rasmussen, K.K, Teilum, K, Kulahin, N, Clausen, O, Berezin, V, Bock, E, Walmod, P.S, Poulsen, F.M. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second Fibronectin type-III module of NCAM2
To be Published
|
|
4NMT
 
 | |
2KRC
 
 | | Solution structure of the N-terminal domain of Bacillus subtilis delta subunit of RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit delta | | Authors: | Motackova, V, Sanderova, H, Zidek, L, Novacek, J, Padrta, P, Svenkova, A, Jonak, J, Krasny, L, Sklenar, V. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of Bacillus subtilis delta subunit of RNA polymerase and its classification based on structural homologs
Proteins, 78, 2010
|
|
