3PG0
 
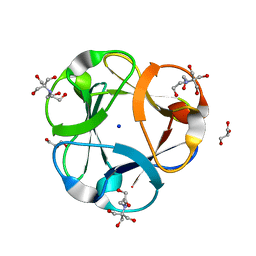 | | Crystal structure of designed 3-fold symmetric protein, ThreeFoil | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SODIUM ION, ... | | Authors: | Lobsanov, Y.D, Broom, A, Howell, P.L, Rose, D.R, Meiering, E.M. | | Deposit date: | 2010-10-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Modular evolution and the origins of symmetry: reconstruction of a three-fold symmetric globular protein.
Structure, 20, 2012
|
|
5RU4
 
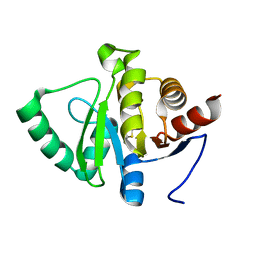 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000001688638 | | Descriptor: | 2-methyl-1,3-thiazole-5-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RUK
 
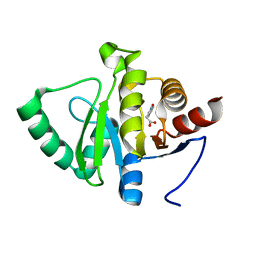 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000161692 | | Descriptor: | 2-(1,2-benzoxazol-3-yl)ethanoic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3H8Z
 
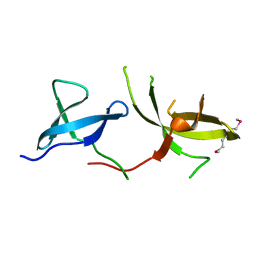 | | The Crystal Structure of the Tudor Domains from FXR2 | | Descriptor: | Fragile X mental retardation syndrome-related protein 2 | | Authors: | Amaya, M.F, Dong, A, Adams-Cioaba, M.A, Guo, Y, MacKenzie, F, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-29 | | Release date: | 2009-06-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Studies of the Tandem Tudor Domains of Fragile X Mental Retardation Related Proteins FXR1 and FXR2.
Plos One, 5, 2010
|
|
3GYT
 
 | | Nuclear receptor DAF-12 from parasitic nematode Strongyloides stercoralis in complex with its physiological ligand dafachronic acid delta 4 | | Descriptor: | (14beta,17alpha,25R)-3-oxocholest-4-en-26-oic acid, Nuclear hormone receptor of the steroid/thyroid hormone receptors superfamily, SRC1 | | Authors: | Zhou, X.E, Wang, Z, Suino-Powell, K, Motola, D.L, Conneely, A, Ogata, C, Sharma, K.K, Auchus, R.J, Kliewer, S.A, Xu, H.E, Mangelsdorf, D.J. | | Deposit date: | 2009-04-05 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the nuclear receptor DAF-12 as a therapeutic target in parasitic nematodes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5RV1
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000251609 | | Descriptor: | Non-structural protein 3, trifluoroacetic acid | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVJ
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000001612349 | | Descriptor: | 6-amino-2H-chromen-2-one, Non-structural protein 3 | | Authors: | Correy, G.C, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-10-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3PL2
 
 | |
4LXK
 
 | | Crystal Structure of Human Beta Secretase in Complex with compound 11d | | Descriptor: | (1R,3S,4S,5R)-3-(4-amino-3-fluoro-5-{[(2R)-1,1,1-trifluoro-3-methoxypropan-2-yl]oxy}benzyl)-5-[(3-tert-butylbenzyl)amino]tetrahydro-2H-thiopyran-4-ol 1-oxide, Beta-secretase 1, GLYCEROL | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of cyclic sulfoxide hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2H70
 
 | | Crystal Structure of Thioredoxin Mutant D9E in Hexagonal (p61) Space Group | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Thioredoxin | | Authors: | Gavira, J.A, Godoy-Ruiz, R, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2006-06-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A stability pattern of protein hydrophobic mutations that reflects evolutionary structural optimization.
Biophys.J., 89, 2005
|
|
4KTE
 
 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE148, from non-human primate | | Descriptor: | GE148 Heavy Chain Fab, GE148 Light Chain Fab, GLYCEROL, ... | | Authors: | Poulsen, C, Tran, K, Stanfield, R, Wyatt, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-05 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2H18
 
 | | Structure of human ADP-ribosylation factor-like 10B (ARL10B) | | Descriptor: | ADP-ribosylation factor-like protein 8A, GUANOSINE-5'-DIPHOSPHATE, UNKNOWN ATOM OR ION | | Authors: | Atanassova, A, Tempel, W, Dimov, S, Yaniw, D, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-16 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structure of human ADP-ribosylation factor-like 10B (ARL10B)
To be Published
|
|
4KLI
 
 | | DNA polymerase beta matched product complex with Mg2+, 90 s | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Freudenthal, B.D, Beard, W.A, Shock, D.D, Wilson, S.H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Observing a DNA polymerase choose right from wrong.
Cell(Cambridge,Mass.), 154, 2013
|
|
4LZ8
 
 | |
3PPG
 
 | | Crystal structure of the Candida albicans methionine synthase by surface entropy reduction, alanine variant with zinc | | Descriptor: | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, ZINC ION | | Authors: | Ubhi, D, Kavanagh, K, Monzingo, A.F, Robertus, J.D. | | Deposit date: | 2010-11-24 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of Candida albicans methionine synthase determined by employing surface residue mutagenesis.
Arch.Biochem.Biophys., 513, 2011
|
|
2R1Z
 
 | |
4LZW
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with thymidine at 1.29 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ETHANOL, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2013-08-01 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | X-ray structures of uridine phosphorylase from Vibrio cholerae in complexes with uridine, thymidine, uracil, thymine, and phosphate anion: Substrate specificity of bacterial uridine phosphorylases
Crystallography Reports, 2016
|
|
2QUO
 
 | |
2R2E
 
 | |
2HAG
 
 | |
2R2Q
 
 | | Crystal structure of human Gamma-Aminobutyric Acid Receptor-Associated Protein-like 1 (GABARAP1), Isoform CRA_a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Gamma-aminobutyric acid receptor-associated protein-like 1, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Paramanathan, R, Davis, T, Mujib, S, Butler-Cole, C, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-27 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of human Gamma-Aminobutyric Acid Receptor-Associated Protein-like 1 (GABARAP1), Isoform CRA_a.
To be Published
|
|
3PFP
 
 | | Structure of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis in complex with an active site inhibitor | | Descriptor: | (2R)-2,7-bis(phosphonooxy)heptanoic acid, (2S)-2,7-bis(phosphonooxy)heptanoic acid, CHLORIDE ION, ... | | Authors: | Reichau, S, Jiao, W, Walker, S.R, Hutton, R.D, Parker, E.J, Baker, E.N. | | Deposit date: | 2010-10-28 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Potent inhibitors of a shikimate pathway enzyme from Mycobacterium tuberculosis: combining mechanism- and modeling-based design
J.Biol.Chem., 286, 2011
|
|
4KMW
 
 | | Structure of the Y34N MUTANT OF DEHALOPEROXIDASE-HEMOGLOBIN A FROM AMPHITRITE ORNATA WITH 2,4,6-TRICHLOROPHENOL | | Descriptor: | 2,4,6-trichlorophenol, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wang, C, Lovelace, L, Lebioda, L. | | Deposit date: | 2013-05-08 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Complexes of dual-function hemoglobin/dehaloperoxidase with substrate 2,4,6-trichlorophenol are inhibitory and indicate binding of halophenol to compound I.
Biochemistry, 52, 2013
|
|
3H6U
 
 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS1493 at 1.85 A resolution | | Descriptor: | (3S)-3-cyclopentyl-6-methyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, CITRATE ANION, GLUTAMIC ACID, ... | | Authors: | Hald, H, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-04-24 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
3PR8
 
 | | Structure of Glutathione S-transferase(PP0183) from Pseudomonas putida in complex with GSH | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLUTATHIONE, Glutathione S-transferase family protein | | Authors: | Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-11-29 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Glutathione S-transferase(PP0183) from Pseudomonas putida in complex with GSH
To be Published
|
|
