2YXF
 
 | |
2Z6R
 
 | | Crystal structure of Lys49 to Arg mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Krishna Swamy, B.S, Simanshu, D.K, Murthy, M.R.N, Kunishima, N. | | Deposit date: | 2007-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
2ZIC
 
 | | Crystal structure of Streptococcus mutans dextran glucosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Dextran glucosidase, ... | | Authors: | Hondoh, H, Saburi, W, Mori, H, Okuyama, M, Nakada, T, Matsuura, Y, Kimura, A. | | Deposit date: | 2008-02-14 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate recognition mechanism of alpha-1,6-glucosidic linkage hydrolyzing enzyme, dextran glucosidase from Streptococcus mutans.
J.Mol.Biol., 378, 2008
|
|
2ZTN
 
 | | Hepatitis E virus ORF2 (Genotype 3) | | Descriptor: | Capsid protein | | Authors: | Yamashita, T, Unno, H, Mori, Y, Li, T.C, Takeda, N, Matsuura, Y. | | Deposit date: | 2008-10-08 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZGW
 
 | | Crystal Structure Of Biotin Protein Ligase From Pyrococcus Horikoshii Complexed with Adenosine and Biotin, Mutations R48A and K111A | | Descriptor: | ADENOSINE, BIOTIN, biotin--[acetyl-CoA-carboxylase] ligase | | Authors: | Bagautdinov, B, Matsuura, Y, Bagautdinova, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protein biotinylation visualized by a complex structure of biotin protein ligase with a substrate
J.Biol.Chem., 283, 2008
|
|
2ZID
 
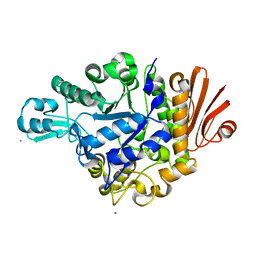 | | Crystal structure of dextran glucosidase E236Q complex with isomaltotriose | | Descriptor: | CALCIUM ION, Dextran glucosidase, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Hondoh, H, Saburi, W, Mori, H, Okuyama, M, Nakada, T, Matsuura, Y, Kimura, A. | | Deposit date: | 2008-02-14 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate recognition mechanism of alpha-1,6-glucosidic linkage hydrolyzing enzyme, dextran glucosidase from Streptococcus mutans.
J.Mol.Biol., 378, 2008
|
|
3A34
 
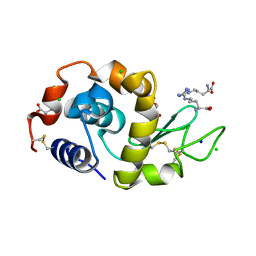 | | Effect of Ariginine on lysozyme | | Descriptor: | ACETATE ION, ARGININE, CHLORIDE ION, ... | | Authors: | Ito, L, Shiraki, K, Matsuura, T, Yamaguchi, H. | | Deposit date: | 2009-06-09 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Implication of Arg bindings on aromatic surfaces of lysozyme for additives that prevent protein aggregation
TO BE PUBLISHED
|
|
1WA5
 
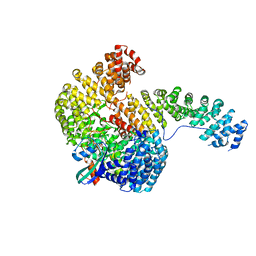 | | Structure of the Cse1:Imp-alpha:RanGTP complex | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, Importin alpha re-exporter, ... | | Authors: | Stewart, M. | | Deposit date: | 2004-10-23 | | Release date: | 2004-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the assembly of a nuclear export complex.
Nature, 432, 2004
|
|
3TPO
 
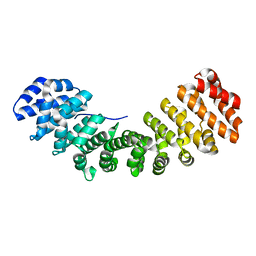 | |
3VOQ
 
 | |
3ULB
 
 | |
3ULC
 
 | |
3VYC
 
 | |
3W3X
 
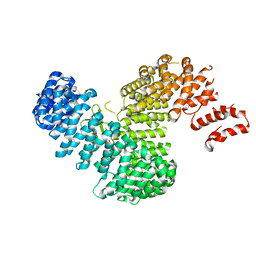 | | Crystal structure of Kap121p bound to Pho4p | | Descriptor: | Importin subunit beta-3, Phosphate system positive regulatory protein PHO4 | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3TPQ
 
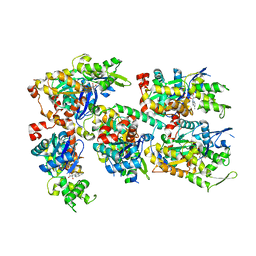 | |
3WUT
 
 | | Structure basis of inactivating cell abscission | | Descriptor: | Centrosomal protein of 55 kDa, GLYCEROL, Inactive serine/threonine-protein kinase TEX14 | | Authors: | Kim, H.J, Matsuura, A, Lee, H.H. | | Deposit date: | 2014-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural and biochemical insights into the role of testis-expressed gene 14 (TEX14) in forming the stable intercellular bridges of germ cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3W3W
 
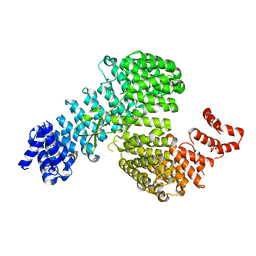 | | Crystal structure of Kap121p bound to Ste12p | | Descriptor: | Importin subunit beta-3, Protein STE12 | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3WUV
 
 | | Structure basis of inactivating cell abscission with chimera peptide 2 | | Descriptor: | Centrosomal protein of 55 kDa, peptide from Programmed cell death 6-interacting protein | | Authors: | Kim, H.J, Matsuura, A, Lee, H.H. | | Deposit date: | 2014-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and biochemical insights into the role of testis-expressed gene 14 (TEX14) in forming the stable intercellular bridges of germ cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3W3Y
 
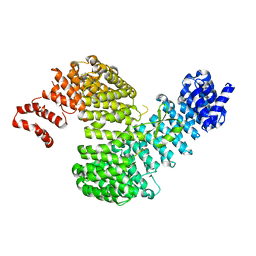 | | Crystal structure of Kap121p bound to Nup53p | | Descriptor: | Importin subunit beta-3, Nucleoporin NUP53 | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3W3T
 
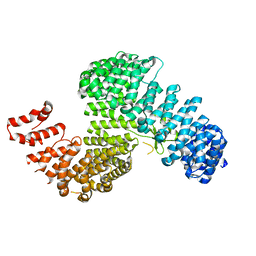 | | Crystal structure of Kap121p | | Descriptor: | Importin subunit beta-3 | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3W3V
 
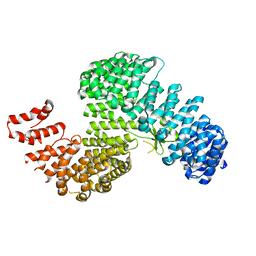 | |
3W3Z
 
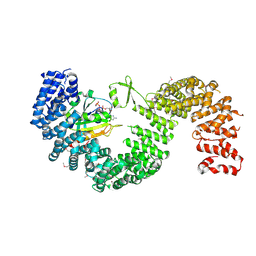 | | Crystal structure of Kap121p bound to RanGTP | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, Importin subunit beta-3, ... | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3W3U
 
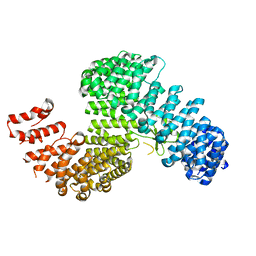 | |
3WUU
 
 | |
3TPM
 
 | |
