4KCM
 
 | | Structure of neuronal nitric oxide synthase heme domain in complex with N-(4-(2-(ethyl(3-(thiophene-2-carboximidamido)benzyl)amino)ethyl)phenyl)thiophene-2-carboximidamide | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, BROMIDE ION, N-(4-{2-[ethyl(3-{[(E)-imino(thiophen-2-yl)methyl]amino}benzyl)amino]ethyl}phenyl)thiophene-2-carboximidamide, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.074 Å) | | Cite: | Potent and Selective Double-Headed Thiophene-2-carboximidamide Inhibitors of Neuronal Nitric Oxide Synthase for the Treatment of Melanoma.
J.Med.Chem., 57, 2014
|
|
7L16
 
 | | Crystal structure of sugar-bound melibiose permease MelB | | Descriptor: | Melibiose carrier protein, dodecyl 6-O-alpha-D-galactopyranosyl-beta-D-glucopyranoside | | Authors: | Guan, L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | X-ray crystallography reveals molecular recognition mechanism for sugar binding in a melibiose transporter MelB.
Commun Biol, 4, 2021
|
|
3I53
 
 | |
4KJW
 
 | |
4H38
 
 | |
4KMY
 
 | | Human folate receptor beta (FOLR2) at neutral pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Folate receptor beta, POTASSIUM ION | | Authors: | Wibowo, A.S, Dann III, C.E. | | Deposit date: | 2013-05-08 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Structures of human folate receptors reveal biological trafficking states and diversity in folate and antifolate recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3HXN
 
 | |
3I0R
 
 | | crystal structure of HIV reverse transcriptase in complex with inhibitor 3 | | Descriptor: | Reverse transcriptase/ribonuclease H, S-{2-[(2-chloro-4-sulfamoylphenyl)amino]-2-oxoethyl} 6-methyl-3,4-dihydroquinoline-1(2H)-carbothioate, p51 RT | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2009-06-25 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Substituted tetrahydroquinolines as potent allosteric inhibitors of reverse transcriptase and its key mutants.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7L17
 
 | | Crystal structure of sugar-bound melibiose permease MelB | | Descriptor: | 4-nitrophenyl alpha-D-galactopyranoside, Melibiose carrier protein | | Authors: | Guan, L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | X-ray crystallography reveals molecular recognition mechanism for sugar binding in a melibiose transporter MelB.
Commun Biol, 4, 2021
|
|
3I2G
 
 | | Cocaine Esterase with mutation G173Q, bound to DTT adduct | | Descriptor: | (4S,5S)-4,5-BIS(MERCAPTOMETHYL)-1,3-DIOXOLAN-2-OL, CHLORIDE ION, Cocaine esterase, ... | | Authors: | Tesmer, J.J.G, Nance, M.R. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of thermostabilizing mutations of cocaine esterase.
Protein Eng.Des.Sel., 23, 2010
|
|
4H3C
 
 | |
3I64
 
 | | Crystal structure of an O-methyltransferase (NcsB1) from neocarzinostatin biosynthesis in complex with S-adenosyl-L-homocysteine (SAH) and 1,4-dihydroxy-2-naphthoic acid (DHN) | | Descriptor: | 1,4-dihydroxy-2-naphthoic acid, GLYCEROL, O-methyltransferase, ... | | Authors: | Cooke, H.A, Bruner, S.D. | | Deposit date: | 2009-07-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis of substrate promiscuity for the SAM-dependent O-methyltransferase NcsB1, involved in the biosynthesis of the enediyne antitumor antibiotic neocarzinostatin.
Biochemistry, 48, 2009
|
|
4H4O
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (RT) in Complex with (E)-3-(3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)- 4-fluorophenoxy)-5-fluorophenyl)acrylonitrile (JLJ506), A Non-nucleoside inhibitor | | Descriptor: | (2E)-3-(3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-5-fluorophenyl)prop-2-enenitrile, Reverse transcriptase/ribonuclease H, Exoribonuclease H, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2012-09-17 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase with Picomolar Inhibitors Reveal Key Interactions for Drug Design.
J.Am.Chem.Soc., 134, 2012
|
|
4LWY
 
 | | L(M196)H,H(M202)L Double Mutant Structure of Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Gabdulkhakov, A.G. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Molecular Dynamic Studies of Reaction Centers Mutants from Rhodobacter sphaeroides and his mutant form L(M196)H+H(M202)L
CRYSTALLOGR REP., 59, 2014
|
|
3HND
 
 | | Crystal structure of human ribonucleotide reductase 1 bound to the effector TTP and substrate GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Fairman, J.W, Wijerathna, S.R, Xu, H, Dealwis, C.G. | | Deposit date: | 2009-05-31 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4M1M
 
 | |
3HR0
 
 | | Crystal structure of Homo sapiens Conserved Oligomeric Golgi subunit 4 | | Descriptor: | CoG4 | | Authors: | Richardson, B.C, Ungar, D, Nakamura, A, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2009-06-08 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for a human glycosylation disorder caused by mutation of the COG4 gene.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4FAM
 
 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 in complex with 3-((3,4-dihydroisoquinolin-2(1H)-yl)sulfonyl)benzoic acid (17) | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,4-dihydroisoquinolin-2(1H)-ylsulfonyl)benzoic acid, Aldo-keto reductase family 1 member C3, ... | | Authors: | Turnbull, A.P, Jamieson, S.M.F, Brooke, D.G, Heinrich, D, Atwell, G.J, Silva, S, Hamilton, E.J, Rigoreau, L.J.M, Trivier, E, Soudy, C, Samlal, S.S, Owen, P.J, Schroeder, E, Raynham, T, Flanagan, J.U, Denny, W.A. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-(3,4-Dihydroisoquinolin-2(1H)-ylsulfonyl)benzoic acids; a New Class of Highly Potent and Selective Inhibitors of the Type 5 17-beta-hydroxysteroid Dehydrogenase AKR1C3
J.Med.Chem., 55, 2012
|
|
4M4A
 
 | | Human Hemoglobin Nitromethane Modified | | Descriptor: | GLYCEROL, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Yi, J, Guan, Y, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2013-08-06 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Degradation of human hemoglobin by organic C-nitroso compounds.
Chem.Commun.(Camb.), 49, 2013
|
|
3I2K
 
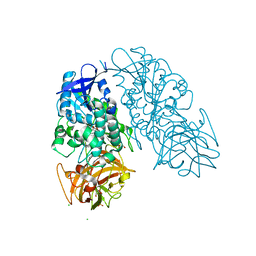 | | Cocaine Esterase, wild type, bound to a DTT adduct | | Descriptor: | (4S,5S)-4,5-BIS(MERCAPTOMETHYL)-1,3-DIOXOLAN-2-OL, CHLORIDE ION, Cocaine esterase, ... | | Authors: | Tesmer, J.J.G, Nance, M.R. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural analysis of thermostabilizing mutations of cocaine esterase.
Protein Eng.Des.Sel., 23, 2010
|
|
3HUS
 
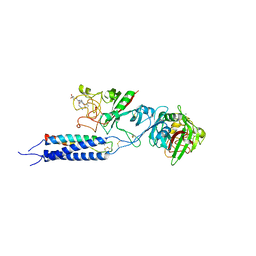 | | Crystal structure of recombinant gamma N308K fibrinogen fragment D with the peptide ligand Gly-Pro-Arg-Pro-amide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fibrinogen alpha chain, ... | | Authors: | Lord, S.T, Bowley, S.R, Okumura, N. | | Deposit date: | 2009-06-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Impaired protofibril formation in fibrinogen gammaN308K is due to altered D:D and "A:a" interactions.
Biochemistry, 48, 2009
|
|
3I4D
 
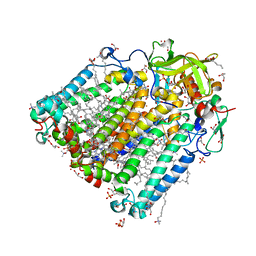 | | Photosynthetic reaction center from rhodobacter sphaeroides 2.4.1 | | Descriptor: | (2R,3S)-heptane-1,2,3-triol, 1,4-DIETHYLENE DIOXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Fujii, R, Adachi, S, Roszak, A.W, Gardiner, A.T, Cogdell, R.J, Isaacs, N.W, Koshihara, S, Hashimoto, H. | | Deposit date: | 2009-07-01 | | Release date: | 2010-12-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the carotenoid bound to the reaction centre from Rhodobacter sphaeroides 2.4.1 revealed by time-resolved X-ray crystallography
To be Published
|
|
7LPX
 
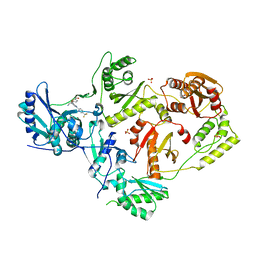 | | Crystal Structure of HIV-1 RT in Complex with NBD-14270 | | Descriptor: | Reverse transcriptase p51, Reverse transcriptase p66, SULFATE ION, ... | | Authors: | Losada, N, Ruiz, F.X, Arnold, E. | | Deposit date: | 2021-02-12 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | HIV-1 gp120 Antagonists Also Inhibit HIV-1 Reverse Transcriptase by Bridging the NNRTI and NRTI Sites.
J.Med.Chem., 64, 2021
|
|
7LQU
 
 | | Crystal Structure of HIV-1 RT in Complex with NBD-14075 | | Descriptor: | Reverse transcriptase p51, Reverse transcriptase p66, SULFATE ION, ... | | Authors: | Losada, N, Ruiz, F.X, Gruber, K, Das, K, Arnold, E. | | Deposit date: | 2021-02-15 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | HIV-1 gp120 Antagonists Also Inhibit HIV-1 Reverse Transcriptase by Bridging the NNRTI and NRTI Sites.
J.Med.Chem., 64, 2021
|
|
7LPW
 
 | | Crystal Structure of HIV-1 RT in Complex with NBD-14189 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Reverse transcriptase p51, ... | | Authors: | Losada, N, Ruiz, F.X, Arnold, E. | | Deposit date: | 2021-02-12 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | HIV-1 gp120 Antagonists Also Inhibit HIV-1 Reverse Transcriptase by Bridging the NNRTI and NRTI Sites.
J.Med.Chem., 64, 2021
|
|
