5DF5
 
 | | The structure of oxidized rat cytochrome c (T28E) at 1.30 angstroms resolution. | | Descriptor: | Cytochrome c, somatic, HEME C, ... | | Authors: | Edwards, B.F.P, Mahapatra, G, Vaishnav, A.A, Brunzelle, J.S, Huttemann, M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | The structure of oxidized rat cytochrome c (T28E) at 1.30 angstroms resolution.
To Be Published
|
|
6TSY
 
 | | Cytochrome c6 from Thermosynechococcus elongatus in a monoclinic space group | | Descriptor: | Cytochrome c6, HEME C, SULFATE ION | | Authors: | Feiler, C.G, Falke, S, Sarrou, I. | | Deposit date: | 2019-12-21 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of native cytochrome c6from Thermosynechococcus elongatus in two different space groups and implications for its oligomerization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4ULV
 
 | | Cytochrome c prime from Shewanella frigidimarina | | Descriptor: | CYTOCHROME C, CLASS II, GLYCEROL, ... | | Authors: | Manole, A.A, Kekilli, D, Dobbin, P.S, Hough, M.A. | | Deposit date: | 2014-05-14 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Conformational Control of the Binding of Diatomic Gases to Cytochrome C'.
J.Biol.Inorg.Chem., 20, 2015
|
|
4W90
 
 | | Crystal structure of Bacillus subtilis cyclic-di-AMP riboswitch ydaO | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A, ... | | Authors: | Jones, C.P, Ferre-D'Amare, A.R. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.118 Å) | | Cite: | Crystal structure of a c-di-AMP riboswitch reveals an internally pseudo-dimeric RNA.
Embo J., 33, 2014
|
|
1U75
 
 | |
6TR1
 
 | | Native cytochrome c6 from Thermosynechococcus elongatus in space group H3 | | Descriptor: | Cytochrome c6, HEME C, SODIUM ION | | Authors: | Falke, S, Feiler, C.G, Sarrou, I. | | Deposit date: | 2019-12-17 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of native cytochrome c6from Thermosynechococcus elongatus in two different space groups and implications for its oligomerization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
2VBJ
 
 | | Molecular basis of human XPC gene recognition and cleavage by engineered homing endonuclease heterodimers | | Descriptor: | 5'-D(*TP*CP*TP*GP*CP*CP*TP*TP*TP*TP *TP*TP*GP*AP*AP*GP*GP*AP*TP*CP*CP*TP*AP*A)-3', 5'-D(*TP*TP*AP*GP*GP*AP*TP*CP*CP*TP *TP*CP*AP*AP*AP*AP*AP*AP*GP*GP*CP*AP*GP*A)-3', CALCIUM ION, ... | | Authors: | Redondo, P, Prieto, J, Munoz, I.G, Alibes, A, Stricher, F, Serrano, L, Arnould, S, Perez, C, Cabaniols, J.P, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2007-09-14 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Basis of Xeroderma Pigmentosum Group C DNA Recognition by Engineered Meganucleases
Nature, 456, 2008
|
|
2VBO
 
 | | Molecular basis of human XPC gene recognition and cleavage by engineered homing endonuclease heterodimers | | Descriptor: | 5'-D(*TP*CP*TP*GP*CP*CP*TP*TP*TP*TP *TP*TP*GP*AP*AP*GP*GP*AP*TP*CP*CP*TP*AP*A)-3', 5'-D(*TP*TP*AP*GP*GP*AP*TP*CP*CP*TP *TP*CP*AP*AP*AP*AP*AP*AP*GP*GP*CP*AP*GP*A)-3', CALCIUM ION, ... | | Authors: | Redondo, P, Prieto, J, Munoz, I.G, Alibes, A, Stricher, F, Serrano, L, Arnould, S, Perez, C, Cabaniols, J.P, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2007-09-14 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of Xeroderma Pigmentosum Group C DNA Recognition by Engineered Meganucleases
Nature, 456, 2008
|
|
6TLF
 
 | | human 14-3-3 sigma isoform in complex with IMP | | Descriptor: | 14-3-3 protein sigma, INOSINIC ACID, SULFATE ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2019-12-02 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of Phosphate-Containing Compounds as New Inhibitors of 14-3-3/c-Abl Protein-Protein Interaction.
Acs Chem.Biol., 15, 2020
|
|
6TM7
 
 | | Human 14-3-3 sigma isoform in complex with PLP | | Descriptor: | 14-3-3 protein sigma, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2019-12-03 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of Phosphate-Containing Compounds as New Inhibitors of 14-3-3/c-Abl Protein-Protein Interaction.
Acs Chem.Biol., 15, 2020
|
|
9R5Y
 
 | | Crystal structure of the C-terminal domain of human TNC in complex with Adhiron 52 | | Descriptor: | Adhiron, Tenascin | | Authors: | Gaule, T.G, Trinh, C, Simmons, K.J, Tomlinson, D.C, Maqbool, A. | | Deposit date: | 2025-05-11 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Targeting Tenascin-C-Toll-like Receptor 4 signalling with Adhiron-derived small molecules - a viable strategy for reducing fibrosis in Systemic Sclerosis
To Be Published
|
|
4V3D
 
 | | The CIDRa domain from HB3var03 PfEMP1 bound to endothelial protein C receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOTHELIAL PROTEIN C RECEPTOR, ... | | Authors: | Lau, C.K.Y, Turner, L, Jespersen, J.S, Lowe, E.D, Petersen, B, Wang, C.W, Petersen, J.E.V, Lusingu, J, Theander, T.G, Lavstsen, T, Higgins, M.K. | | Deposit date: | 2014-10-17 | | Release date: | 2014-12-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Conservation Despite Huge Sequence Diversity Allows Epcr Binding by the Pfemp1 Family Implicated in Severe Childhood Malaria.
Cell Host Microbe., 17, 2015
|
|
6TNT
 
 | | SUMOylated apoAPC/C with repositioned APC2 WHB domain | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Barford, D, Yatskevich, S. | | Deposit date: | 2019-12-10 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular mechanisms of APC/C release from spindle assembly checkpoint inhibition by APC/C SUMOylation.
Cell Rep, 34, 2021
|
|
9MTW
 
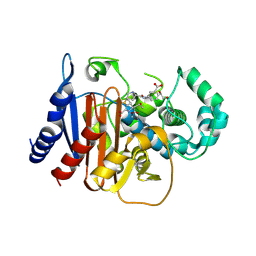 | | Crystal structure of ADC-1-ertapenem complex | | Descriptor: | (2S,3R,4R)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, ... | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2025-01-12 | | Release date: | 2025-04-23 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolution of carbapenemase activity in the class C beta-lactamase ADC-1.
Mbio, 16, 2025
|
|
6LBF
 
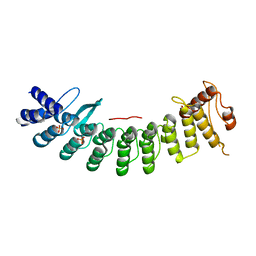 | | Crystal structure of FEM1B | | Descriptor: | Protein fem-1 homolog B, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.252 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
9MTV
 
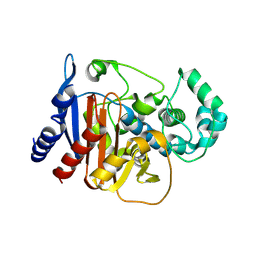 | | Crystal structure of an apo-ADC-1 triple mutant | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, NITRATE ION | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2025-01-12 | | Release date: | 2025-04-23 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of carbapenemase activity in the class C beta-lactamase ADC-1.
Mbio, 16, 2025
|
|
9MTU
 
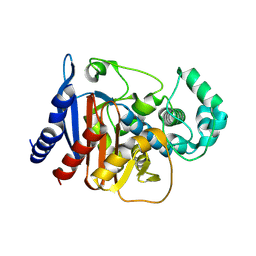 | |
4UIV
 
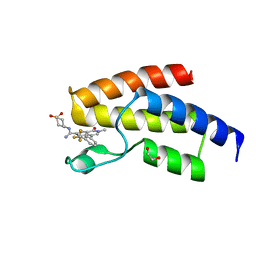 | | BROMODOMAIN OF HUMAN BRD9 WITH N-(1,1-dioxo-1-thian-4-yl)-5-methyl-4- oxo-7-3-(trifluoromethyl)phenyl-4H,5H-thieno-3,2-c-pyridine-2- carboximidamide | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN-CONTAINING PROTEIN 9, N-(1,1-dioxo-1-thian-4-yl)-5-methyl-4-oxo-7-3-(trifluoromethyl)phenyl-4H,5H-thieno-3,2-c-pyridine-2-carboximidamide | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
4UIU
 
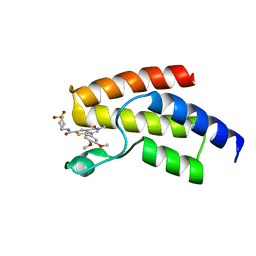 | | BROMODOMAIN OF HUMAN BRD9 WITH 7-(3,4-dimethoxyphenyl)-N-(1,1-dioxo-1- thian-4-yl)-5-methyl-4-oxo-4H,5H-thieno-3,2-c-pyridine-2-carboxamide | | Descriptor: | BROMODOMAIN-CONTAINING PROTEIN 9, N-[1,1-bis(oxidanylidene)thian-4-yl]-7-(3,4-dimethoxyphenyl)-5-methyl-4-oxidanylidene-thieno[3,2-c]pyridine-2-carboxamide | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
4UIY
 
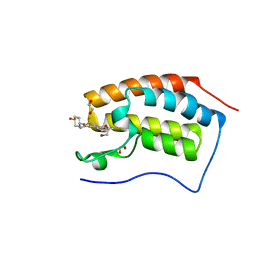 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH N-(1,1-dioxo-1-thian-4-yl)- 5-methyl-4-oxo-7-3-(trifluoromethyl)phenyl-4H,5H-thieno-3,2-c- pyridine-2-carboximidamide | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN-CONTAINING PROTEIN 4, N-[1,1-bis(oxidanylidene)thian-4-yl]-5-methyl-4-oxidanylidene-7-[3-(trifluoromethyl)phenyl]thieno[3,2-c]pyridine-2-carboximidamide | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
2V23
 
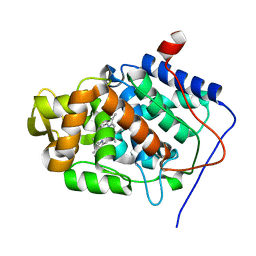 | | Structure of cytochrome c peroxidase mutant N184R Y36A | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Metcalfe, C.L, Macdonald, I.K, Brown, K.A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2007-05-31 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Tuberculosis Prodrug Isoniazid Bound to Activating Peroxidases.
J.Biol.Chem., 283, 2008
|
|
4UIW
 
 | | BROMODOMAIN OF HUMAN BRD9 WITH N-(1,1-dioxo-1-thian-4-yl)-5-ethyl-4- oxo-7-3-(trifluoromethyl)phenyl-4H,5H-thieno-3,2-c-pyridine-2- carboximidamide | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN-CONTAINING PROTEIN 9, N'-[1,1-bis(oxidanylidene)thian-4-yl]-5-ethyl-4-oxidanylidene-7-[3-(trifluoromethyl)phenyl]thieno[3,2-c]pyridine-2-carboximidamide | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
2V2E
 
 | | Structure of isoniazid (INH) bound to cytochrome c peroxidase mutant N184R Y36A | | Descriptor: | 4-(DIAZENYLCARBONYL)PYRIDINE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Metcalfe, C.L, Macdonald, I.K, Brown, K.A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2007-06-05 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Tuberculosis Prodrug Isoniazid Bound to Activating Peroxidases.
J.Biol.Chem., 283, 2008
|
|
4UIT
 
 | | BROMODOMAIN OF HUMAN BRD9 WITH 7-(3,4-dimethoxyphenyl)-2-(4- methanesulfonylpiperazine-1-carbonyl)-5-methyl-4H,5H-thieno-3,2-c- pyridin-4-one | | Descriptor: | 7-(3,4-dimethoxyphenyl)-5-methyl-2-(4-methylsulfonylpiperazin-1-yl)carbonyl-thieno[3,2-c]pyridin-4-one, BROMODOMAIN-CONTAINING PROTEIN 9 | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
4UIZ
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH 7-(3,4-dimethoxyphenyl)-2-(4-methanesulfonylpiperazine-1-carbonyl)-5-methyl-4H,5H-thieno-3,2-c- pyridin-4-one | | Descriptor: | 1,2-ETHANEDIOL, 7-(3,4-dimethoxyphenyl)-5-methyl-2-(4-methylsulfonylpiperazin-1-yl)carbonyl-thieno[3,2-c]pyridin-4-one, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
