4YC6
 
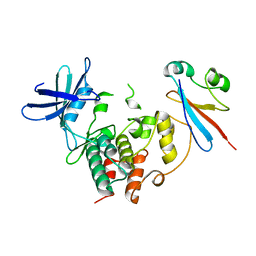 | | CDK1/CKS1 | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 1 | | Authors: | Brown, N.R, Korolchuk, S, Martin, M.P, Stanley, W, Moukhametzianov, R, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2015-02-19 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CDK1 structures reveal conserved and unique features of the essential cell cycle CDK.
Nat Commun, 6, 2015
|
|
5L8Y
 
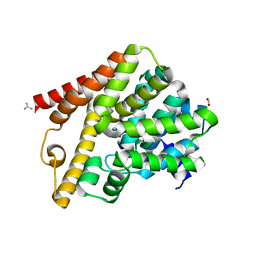 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-937 | | Descriptor: | 2-[(4-{5-[(4aR,8aS)-3-cycloheptyl-4-oxo-3,4,4a,5,8,8a-hexahydrophthalazin-1-yl]-2-methoxyphenyl}phenyl)formamido]-N-(2-hydroxyethyl)acetamide, FORMIC ACID, GLYCEROL, ... | | Authors: | Singh, A.K, Anthonyrajah, E.S, Brown, D.G. | | Deposit date: | 2016-06-08 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
6WAI
 
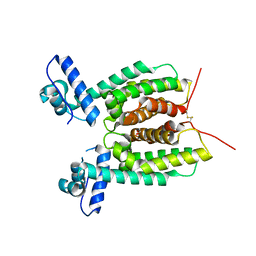 | | Crystal Structure of SmcR N142D from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, LuxR family transcriptional regulator, SULFATE ION | | Authors: | Newman, J.D, Russell, M.M, Gonzalez-Gutierrez, G, van Kessel, J.C. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.583 Å) | | Cite: | The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Nucleic Acids Res., 49, 2021
|
|
5KM1
 
 | |
5L97
 
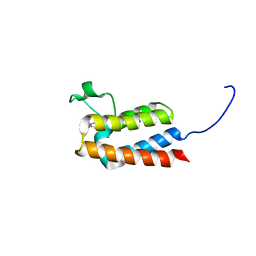 | |
4YDD
 
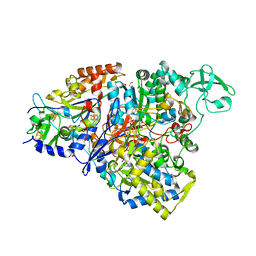 | | Crystal structure of the perchlorate reductase PcrAB from Azospira suillum PS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tsai, C.-L, Youngblut, M.D, Tainer, J.A. | | Deposit date: | 2015-02-21 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
6WBJ
 
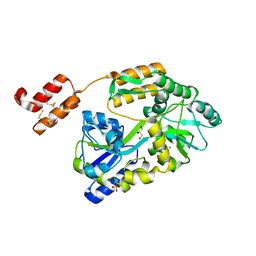 | | High resolution crystal structure of mRECK(CC4) in fusion with engineered MBP | | Descriptor: | GLYCEROL, Maltodextrin-binding protein,Reversion-inducing cysteine-rich protein with Kazal motifs fusion, SULFATE ION, ... | | Authors: | Chang, T.H, Hsieh, F.L, Gabelli, S.B, Nathans, J. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structure of the RECK CC domain, an evolutionary anomaly.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8FUA
 
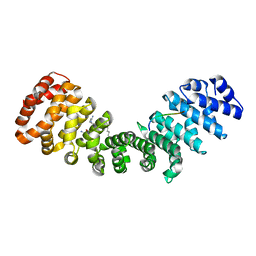 | | Crystal structure of mouse Importin alpha in complex with Hendra virus matrix protein NLS1 | | Descriptor: | Importin subunit alpha-1, Matrix protein | | Authors: | Donnelly, C.M, Basler, C.F, Scott, C, Forwood, J.K. | | Deposit date: | 2023-01-17 | | Release date: | 2023-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Henipavirus Matrix Protein Employs a Non-Classical Nuclear Localization Signal Binding Mechanism.
Viruses, 15, 2023
|
|
5KMA
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside D-Trp phosphoramidate substrate complex | | Descriptor: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-ethyl-phosphonamidic acid | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
8F94
 
 | |
6VJE
 
 | | Crystal structure of Pseudomonas aeruginosa penicillin-binding protein 3 (PBP3) complexed with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, CHLORIDE ION, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2020-01-15 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Insights into Ceftobiprole Inhibition of Pseudomonas aeruginosa Penicillin-Binding Protein 3.
Antimicrob.Agents Chemother., 64, 2020
|
|
5LAF
 
 | |
8FB3
 
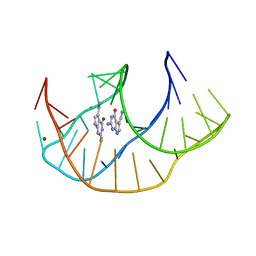 | |
6VKC
 
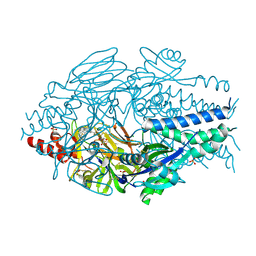 | | Crystal Structure of Inhibitor JNJ-36811054 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 3-{[5-chloro-1-(4,4,4-trifluorobutyl)-1H-imidazo[4,5-b]pyridin-2-yl]methyl}-1-cyclopropyl-1,3-dihydro-2H-imidazo[4,5-c]pyridin-2-one, CHLORIDE ION, D(-)-TARTARIC ACID, ... | | Authors: | McLellan, J.S. | | Deposit date: | 2020-01-20 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 3-({5-Chloro-1-[3-(methylsulfonyl)propyl]-1H-indol-2-yl}methyl)-1-(2,2,2-trifluoroethyl)-1,3-dihydro-2H-imidazo[4,5-c]pyridin-2-one (JNJ-53718678), a Potent and Orally Bioavailable Fusion Inhibitor of Respiratory Syncytial Virus.
J.Med.Chem., 63, 2020
|
|
8I4J
 
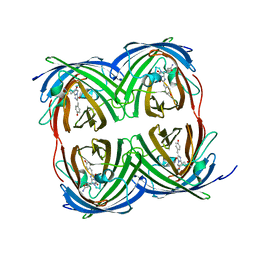 | |
4YOO
 
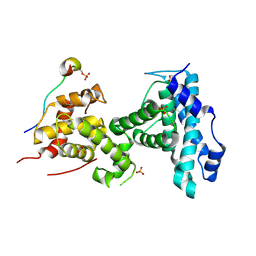 | | p107 pocket domain in complex with LIN52 P29A peptide | | Descriptor: | LIN52 peptide, Retinoblastoma-like protein 1,Retinoblastoma-like protein 1, SULFATE ION | | Authors: | Guiley, K.Z, Liban, T.J, Felthousen, J.G, Ramanan, P, Tripathi, S, Litovchick, L, Rubin, S.M. | | Deposit date: | 2015-03-12 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural mechanisms of DREAM complex assembly and regulation.
Genes Dev., 29, 2015
|
|
4YP0
 
 | |
5KM3
 
 | |
6W0C
 
 | | Open-gate KcsA soaked in 4 mM BaCl2 | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.556 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
8IIY
 
 | |
6VMY
 
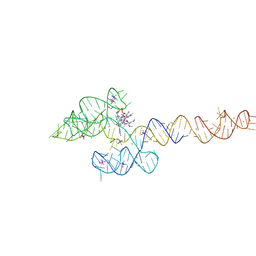 | | Structure of the B. subtilis cobalamin riboswitch | | Descriptor: | Adenosylcobalamin, B. subtilis cobalamin riboswitch, COBALT HEXAMMINE(III), ... | | Authors: | Chan, C.W, Mondragon, A. | | Deposit date: | 2020-01-28 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of an atypical cobalamin riboswitch reveals RNA structural adaptability as basis for promiscuous ligand binding.
Nucleic Acids Res., 48, 2020
|
|
6W0L
 
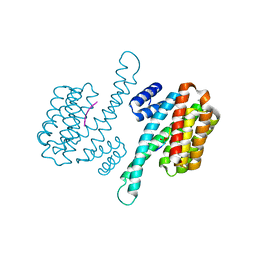 | | Henipavirus W protein interacts with 14-3-3 to modulate host gene expression | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, Phosphorylated W peptide | | Authors: | Edwards, M, Hoad, M, Tsimbalyuk, S, Menicucci, A, Messaoudi, I, Forwood, J, Basler, C. | | Deposit date: | 2020-03-01 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Henipavirus W Proteins Interact with 14-3-3 To Modulate Host Gene Expression.
J.Virol., 94, 2020
|
|
5KOR
 
 | | Arabidopsis thaliana fucosyltransferase 1 (FUT1) in complex with GDP and a xylo-oligossacharide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Rocha, J, de Sanctis, D, Breton, C. | | Deposit date: | 2016-07-01 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Arabidopsis thaliana FUT1 Reveals a Variant of the GT-B Class Fold and Provides Insight into Xyloglucan Fucosylation.
Plant Cell, 28, 2016
|
|
4YRE
 
 | |
6W0S
 
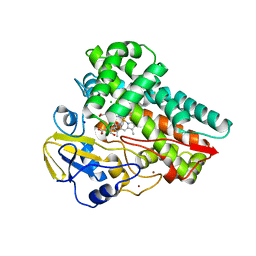 | | Crystal structure of substrate free cytochrome P450 NasF5053 from Streptomyces sp. NRRL F-5053 | | Descriptor: | BROMIDE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-03-02 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
