8FI5
 
 | |
8HUN
 
 | | X-ray structure of human PPAR alpha ligand binding domain-seladelpar co-crystals obtained by cross-seeding | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, Seladelpar | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Iino, S, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUP
 
 | | X-ray structure of human PPAR gamma ligand binding domain-seladelpar-SRC1 coactivator peptide co-crystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, Isoform 1 of Peroxisome proliferator-activated receptor gamma, Seladelpar | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8F9Y
 
 | | SAL1 from Arabidopsis thaliana | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, SAL1 phosphatase | | Authors: | Frkic, R.L, Kaczmarski, J.A, Tan, L, Jackson, C.J. | | Deposit date: | 2022-11-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sensing and signaling of oxidative stress in chloroplasts by inactivation of the SAL1 phosphoadenosine phosphatase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8I50
 
 | | Crystal structure of DNA octamer containing GuNA[Me,Me] | | Descriptor: | DNA (5'-D(*GP*(OIQ)P*GP*(BRU)P*AP*CP*AP*C)-3') | | Authors: | Aoyama, H, Obika, S, Yamaguchi, T. | | Deposit date: | 2023-01-21 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Mechanism of the extremely high duplex-forming ability of oligonucleotides modified with N-tert-butylguanidine- or N-tert-butyl-N'- methylguanidine-bridged nucleic acids.
Nucleic Acids Res., 51, 2023
|
|
6W3B
 
 | | Structure of apo unphosphorylated IRE1 | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Wallweber, H, Mortara, K, Ferri, E, Rudolph, J, Wang, W. | | Deposit date: | 2020-03-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
5KYC
 
 | |
5KYF
 
 | |
6W3Y
 
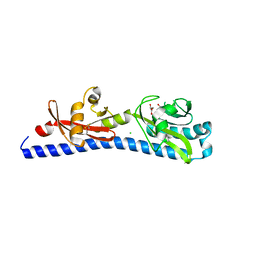 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-alanine | | Descriptor: | ALANINE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
8FT7
 
 | |
8FUQ
 
 | |
6W68
 
 | |
4YEM
 
 | | Carboplatin binding to HEWL in NaBr crystallisation conditions studied at an X-ray wavelength of 0.9163A - new refinement | | Descriptor: | ACETATE ION, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8FUC
 
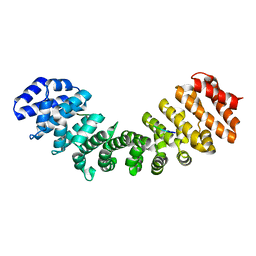 | | Crystal structure of mouse Importin alpha in complex with Hendra virus matrix protein minor site NLS2 | | Descriptor: | Contaminant peptide KKLARE, Importin subunit alpha-1, Matrix protein | | Authors: | Donnelly, C.M, Basler, C.F, Scott, C, Forwood, J.K. | | Deposit date: | 2023-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Henipavirus Matrix Protein Employs a Non-Classical Nuclear Localization Signal Binding Mechanism.
Viruses, 15, 2023
|
|
5KZX
 
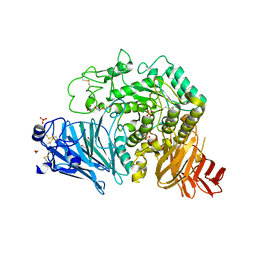 | | Crystal structure of human GAA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Deming, D.T, Garman, S.C. | | Deposit date: | 2016-07-25 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of human GAA: structural basis of Pompe disease
To be published
|
|
4YEX
 
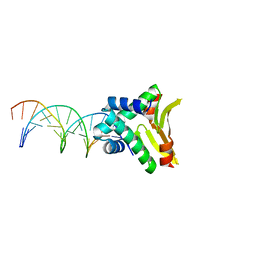 | | HUaa-19bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4Y8V
 
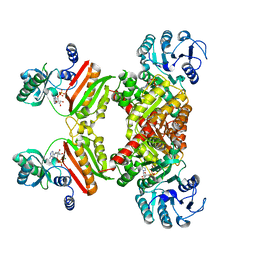 | |
8FZM
 
 | |
6WBH
 
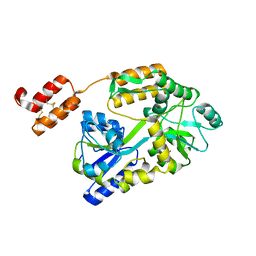 | | Crystal structure of mRECK(CC4) in fusion with engineered MBP at medium resolution | | Descriptor: | CHLORIDE ION, Maltodextrin-binding protein,Reversion-inducing cysteine-rich protein with Kazal motifs fusion, ZINC ION, ... | | Authors: | Chang, T.H, Hsieh, F.L, Gabelli, S.B, Nathans, J. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Structure of the RECK CC domain, an evolutionary anomaly.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8FCD
 
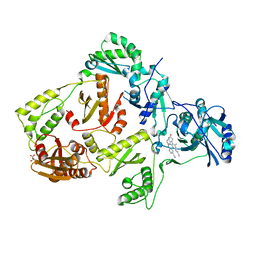 | | HIV-1 Reverse Transcriptase in complex with 6-membered bicyclic core NNRTI | | Descriptor: | 4-[(8-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}-6-oxo-5,6,7,8-tetrahydropteridin-2-yl)amino]benzonitrile, L(+)-TARTARIC ACID, p51 RT, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2022-12-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design and Synthesis of Novel HIV-1 NNRTIs with Bicyclic Cores and with Improved Physicochemical Properties.
J.Med.Chem., 66, 2023
|
|
6VS8
 
 | |
5L2Q
 
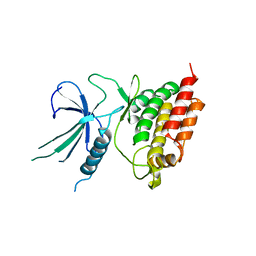 | |
8FCC
 
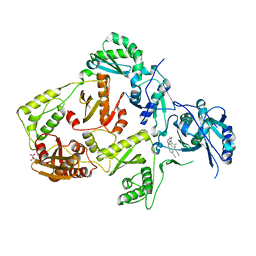 | | HIV-1 Reverse Transcriptase in complex with 5-membered bicyclic core NNRTI | | Descriptor: | 4-[(9-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}-8-oxo-8,9-dihydro-7H-purin-2-yl)amino]benzonitrile, L(+)-TARTARIC ACID, p51 RT, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2022-12-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design and Synthesis of Novel HIV-1 NNRTIs with Bicyclic Cores and with Improved Physicochemical Properties.
J.Med.Chem., 66, 2023
|
|
5L3Y
 
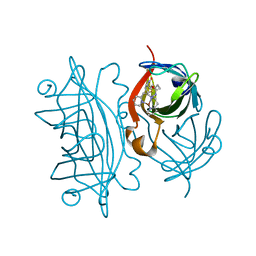 | | Designed Artificial Cupredoxins | | Descriptor: | Streptavidin, [CuII(biot-et-dpea)]2+ | | Authors: | Mann, S.I, Heinisch, T, Weitz, A.C, Hendrich, M.R, Ward, T.R, Borovik, A.S. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Modular Artificial Cupredoxins.
J.Am.Chem.Soc., 138, 2016
|
|
6VUH
 
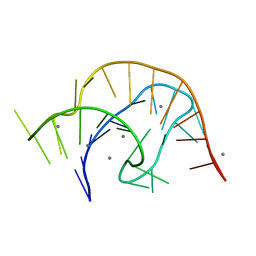 | | APO PreQ1 riboswitch aptamer grown in Mn2+ | | Descriptor: | MANGANESE (II) ION, PREQ1 RIBOSWITCH | | Authors: | Jenkins, J.L, Wedekind, J.E. | | Deposit date: | 2020-02-15 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Analysis of a preQ1-I riboswitch in effector-free and bound states reveals a metabolite-programmed nucleobase-stacking spine that controls gene regulation.
Nucleic Acids Res., 48, 2020
|
|
