4HYS
 
 | |
2QFJ
 
 | | Crystal Structure of First Two RRM Domains of FIR Bound to ssDNA from a Portion of FUSE | | Descriptor: | DNA (5'-D(*DTP*DCP*DGP*DGP*DGP*DAP*DTP*DTP*DTP*DTP*DTP*DTP*DAP*DTP*DTP*DTP*DTP*DGP*DTP*DGP*DTP*DTP*DAP*DTP*DT)-3'), FBP-interacting repressor | | Authors: | Crichlow, G.V, Yang, Y, Fan, C, Lolis, E, Braddock, D. | | Deposit date: | 2007-06-27 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dimerization of FIR upon FUSE DNA binding suggests a mechanism of c-myc inhibition
EMBO J., 27, 2007
|
|
2D25
 
 | | C-C-A-G-G-C-M5C-T-G-G; HELICAL FINE STRUCTURE, HYDRATION, AND COMPARISON WITH C-C-A-G-G-C-C-T-G-G | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*CP*(5CM)P*TP*GP*G)-3'), MAGNESIUM ION | | Authors: | Heinemann, U, Hahn, M. | | Deposit date: | 1991-04-23 | | Release date: | 1991-04-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | C-C-A-G-G-C-m5C-T-G-G. Helical fine structure, hydration, and comparison with C-C-A-G-G-C-C-T-G-G.
J.Biol.Chem., 267, 1992
|
|
7N21
 
 | | NMR structure of AnIB-OH | | Descriptor: | Alpha-conotoxin AnIB | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N22
 
 | | NMR structure of AnIB[Y(SO3)16Y]-NH2 | | Descriptor: | Alpha-conotoxin AnIB | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N20
 
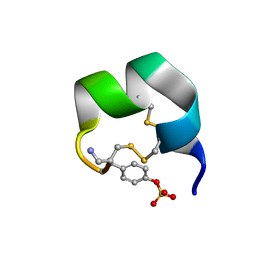 | | NMR structure of native AnIB | | Descriptor: | Alpha-conotoxin AnIB | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N23
 
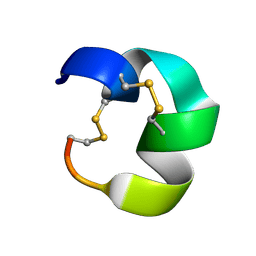 | | NMR structure of AnIB[Y(SO3)16Y]-OH | | Descriptor: | Alpha-conotoxin AnIB | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N0T
 
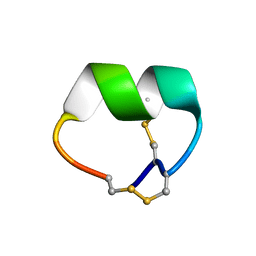 | | NMR structure of EpI[Y(SO)315Y]-OH | | Descriptor: | Alpha-conotoxin EpI | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
4WIW
 
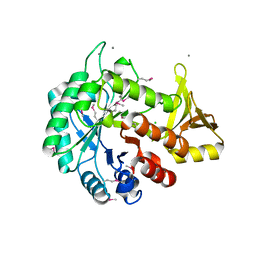 | | Crystal structure of C-terminal domain of putative chitinase from Desulfitobacterium hafniense DCB-2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-26 | | Release date: | 2014-10-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.637 Å) | | Cite: | Crystal structure of C-terminal domain of putative chitinase from Desulfitobacterium hafniense DCB-2
To Be Published
|
|
4LKU
 
 | |
1KJS
 
 | | NMR SOLUTION STRUCTURE OF C5A AT PH 5.2, 303K, 20 STRUCTURES | | Descriptor: | C5A | | Authors: | Zhang, X, Boyar, W, Toth, M, Wennogle, L, Gonnella, N.C. | | Deposit date: | 1997-01-09 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structural definition of the C5a C terminus by two-dimensional nuclear magnetic resonance spectroscopy.
Proteins, 28, 1997
|
|
3S0A
 
 | | Apis mellifera OBP14, native apo-protein | | Descriptor: | OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
3CJ4
 
 | |
3CSO
 
 | | HCV Polymerase in complex with a 1,5 Benzodiazepine inhibitor | | Descriptor: | (11S)-10-acetyl-11-[4-(benzyloxy)-3-chlorophenyl]-3,3-dimethyl-2,3,4,5,10,11-hexahydro-1H-dibenzo[b,e][1,4]diazepin-1-one, RNA-directed RNA polymerase | | Authors: | Nyanguile, O. | | Deposit date: | 2008-04-10 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | 1,5-benzodiazepines, a novel class of hepatitis C virus polymerase nonnucleoside inhibitors.
Antimicrob.Agents Chemother., 52, 2008
|
|
3D1D
 
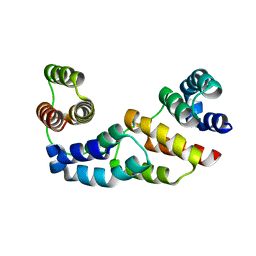 | | Hexagonal crystal structure of Tas3 C-terminal alpha motif | | Descriptor: | RNA-induced transcriptional silencing complex protein tas3 | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2008-05-05 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An alpha motif at Tas3 C terminus mediates RITS cis spreading and promotes heterochromatic gene silencing.
Mol.Cell, 34, 2009
|
|
3D1B
 
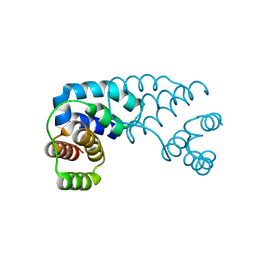 | | Tetragonal crystal structure of Tas3 C-terminal alpha motif | | Descriptor: | RNA-induced transcriptional silencing complex protein tas3 | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2008-05-05 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An alpha motif at Tas3 C terminus mediates RITS cis spreading and promotes heterochromatic gene silencing.
Mol.Cell, 34, 2009
|
|
3DOB
 
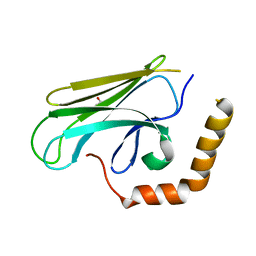 | | Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans. | | Descriptor: | BETA-MERCAPTOETHANOL, Heat shock 70 kDa protein F44E5.5 | | Authors: | Osipiuk, J, Hatzos, C, Gu, M, Zhang, R, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-03 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | X-ray crystal structure of Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans.
To be Published
|
|
2I1U
 
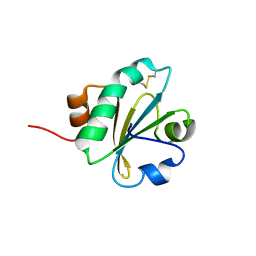 | |
3D1I
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with nitrite | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-05-06 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens
J.Mol.Biol., 389, 2009
|
|
8RVC
 
 | | Crystal structure of alpha keto acid C-methyl-transferases MrsA bound to ketoarginine | | Descriptor: | 1,2-ETHANEDIOL, 2-ketoarginine methyltransferase, 5-[(diaminomethylidene)amino]-2-oxopentanoic acid, ... | | Authors: | Gerhardt, S, Kemper, F, Andexer, J.N. | | Deposit date: | 2024-02-01 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structures and Protein Engineering of the alpha-Keto Acid C-Methyltransferases SgvM and MrsA for Rational Substrate Transfer.
Chembiochem, 25, 2024
|
|
8RWM
 
 | |
8RWW
 
 | |
8RXG
 
 | |
8RVS
 
 | |
8RXF
 
 | |
