1PH7
 
 | |
6CR1
 
 | | adalimumab EFab | | Descriptor: | DI(HYDROXYETHYL)ETHER, Heavy chain of adalimumab EFab (VH-IgE CH2), Light chain of adalimumab EFab (VL-IgE CH2), ... | | Authors: | Arndt, J.W. | | Deposit date: | 2018-03-16 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | EFab domain substitution as a solution to the light-chain pairing problem of bispecific antibodies.
MAbs, 10, 2018
|
|
1MM0
 
 | | Solution structure of termicin, an antimicrobial peptide from the termite Pseudacanthotermes spiniger | | Descriptor: | Termicin | | Authors: | Da Silva, P, Jouvensal, L, Lamberty, M, Bulet, P, Caille, A, Vovelle, F. | | Deposit date: | 2002-09-02 | | Release date: | 2003-05-13 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of termicin, an antimicrobial peptide from the termite Pseudacanthotermes spiniger
PROTEIN SCI., 12, 2003
|
|
5YNZ
 
 | | Crystal structure of the dihydroorotase domain (K1556A) of human CAD | | Descriptor: | CAD protein, ZINC ION | | Authors: | Huang, Y.H, Chen, K.L, Cheng, J.H, Huang, C.Y. | | Deposit date: | 2017-10-26 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.774 Å) | | Cite: | Crystal structures of monometallic dihydropyrimidinase and the human dihydroorotase domain K1556A mutant reveal no lysine carbamylation within the active site
Biochem. Biophys. Res. Commun., 505, 2018
|
|
1UNO
 
 | | Crystal structure of a d,l-alternating peptide | | Descriptor: | H-(L-TYR-D-TYR)4-LYS-OH | | Authors: | Alexopoulos, E, Kuesel, A, Uson, I, Diederichsen, U, Sheldrick, G.M. | | Deposit date: | 2003-09-11 | | Release date: | 2004-09-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Solution and Structure of an Alternating D,L-Peptide
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5Z09
 
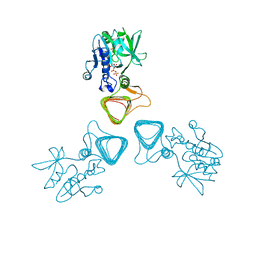 | | ST0452(Y97N)-UTP binding form | | Descriptor: | Dual sugar-1-phosphate nucleotidylyltransferase, URIDINE 5'-TRIPHOSPHATE | | Authors: | Honda, Y, Nakano, S, Ito, S, Dadashipour, M, Zhang, Z, Kawarabayasi, Y. | | Deposit date: | 2017-12-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Improvement of ST0452N-Acetylglucosamine-1-Phosphate Uridyltransferase Activity by the Cooperative Effect of Two Single Mutations Identified through Structure-Based Protein Engineering
Appl. Environ. Microbiol., 84, 2018
|
|
1M3V
 
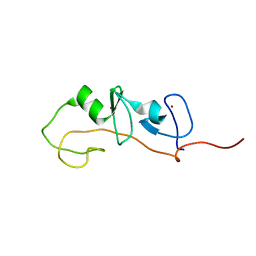 | | FLIN4: Fusion of the LIM binding domain of Ldb1 and the N-terminal LIM domain of LMO4 | | Descriptor: | ZINC ION, fusion of the LIM interacting domain of ldb1 and the N-terminal LIM domain of LMO4 | | Authors: | Deane, J.E, Mackay, J.P, Kwan, A.H.Y, Sum, E.Y, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2002-06-30 | | Release date: | 2003-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4
EMBO J., 22, 2003
|
|
1QI9
 
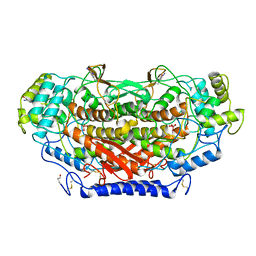 | | X-RAY SIRAS STRUCTURE DETERMINATION OF A VANADIUM-DEPENDENT HALOPEROXIDASE FROM ASCOPHYLLUM NODOSUM AT 2.0 A RESOLUTION | | Descriptor: | VANADATE ION, Vanadium-dependent bromoperoxidase | | Authors: | Weyand, M, Hecht, H.-J, Kiess, M, Liaud, M.F, Vilter, H, Schomburg, D. | | Deposit date: | 1999-06-10 | | Release date: | 2000-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray structure determination of a vanadium-dependent haloperoxidase from Ascophyllum nodosum at 2.0 A resolution.
J.Mol.Biol., 293, 1999
|
|
3I3Z
 
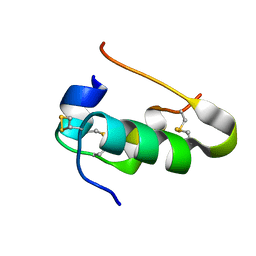 | | Human insulin | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Timofeev, V.I, Bezuglov, V.V, Miroshnikov, K.A, Cuprov-Netochin, R.N, Samigina, V.R, Kuranova, I.P. | | Deposit date: | 2009-07-01 | | Release date: | 2010-01-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray investigation of gene-engineered human insulin crystallized from a solution containing polysialic acid.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1MZT
 
 | |
1L9Q
 
 | |
1L9R
 
 | |
1L9T
 
 | |
7XAX
 
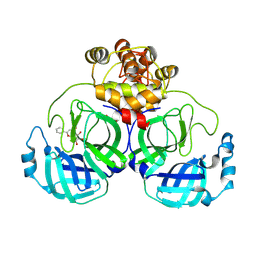 | | Crystal structure of SARS coronavirus main protease in complex with Baicalei | | Descriptor: | 3C-like proteinase nsp5, 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one | | Authors: | Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2022-03-19 | | Release date: | 2023-03-22 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of SARS-CoV 3C-like protease with baicalein.
Biochem.Biophys.Res.Commun., 611, 2022
|
|
1QZ9
 
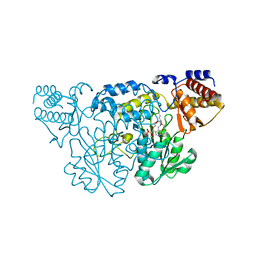 | | The Three Dimensional Structure of Kynureninase from Pseudomonas fluorescens | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECANE, CHLORIDE ION, KYNURENINASE, ... | | Authors: | Momany, C, Levdikov, V, Blagova, L, Lima, S, Phillips, R.S. | | Deposit date: | 2003-09-16 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-Dimensional Structure of Kynureninase from Pseudomonas fluorescens.
Biochemistry, 43, 2004
|
|
1L9O
 
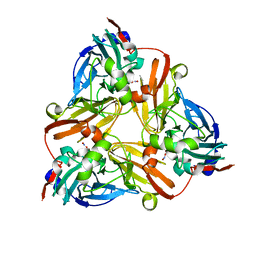 | |
1R48
 
 | | Solution structure of the C-terminal cytoplasmic domain residues 468-497 of Escherichia coli protein ProP | | Descriptor: | Proline/betaine transporter | | Authors: | Zoetewey, D.L, Tripet, B.P, Kutateladze, T.G, Overduin, M.J, Wood, J.M, Hodges, R.S. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Antiparallel Coiled-coil Domain from Escherichia coli Osmosensor ProP.
J.Mol.Biol., 334, 2003
|
|
1HLY
 
 | | SOLUTION STRUCTURE OF HONGOTOXIN 1 | | Descriptor: | HONGOTOXIN 1 | | Authors: | Pragl, B, Koschak, A, Trieb, M, Obermair, G, Kaufmann, W.A, Gerster, U, Blanc, E, Hahn, C, Prinz, H, Schutz, G, Darbon, H, Gruber, H.J, Knaus, H.G. | | Deposit date: | 2000-12-04 | | Release date: | 2003-09-30 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Synthesis, characterization, and application of cy-dye- and alexa-dye-labeled hongotoxin(1) analogues. The
first high affinity fluorescence probes for voltage-gated K+ channels.
BIOCONJUG.CHEM., 13, 2002
|
|
6EZW
 
 | | Crystal structure of a llama VHH antibody BCD090-M2 against human ErbB3 in space group C2 | | Descriptor: | VHH antibody BCD090-M2 | | Authors: | Eliseev, I.E, Yudenko, A.N, Vysochinskaya, V.V, Svirina, A.A, Evstratyeva, A.V, Drozhzhachih, M.S, Krendeleva, E.A, Vladimirova, A.K, Nemankin, T.A, Ekimova, V.M, Ulitin, A.B, Lomovskaya, M.I, Yakovlev, P.A, Moiseenko, F.V, Chakchir, O.B. | | Deposit date: | 2017-11-16 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59842849 Å) | | Cite: | Crystal structures of a llama VHH antibody BCD090-M2 targeting human ErbB3 receptor.
F1000Res, 7, 2018
|
|
1L9P
 
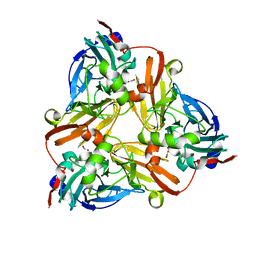 | |
6BCB
 
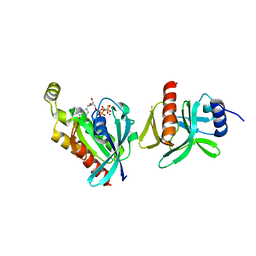 | |
1LKA
 
 | | Porcine Pancreatic Elastase/Ca-Complex | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Weiss, M.S, Panjikar, S, Nowak, E, Tucker, P.A. | | Deposit date: | 2002-04-24 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal binding to porcine pancreatic elastase: calcium or not calcium.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6D71
 
 | | Crystal Structure of the Human Miro1 N-terminal GTPase bound to GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial Rho GTPase 1 | | Authors: | Smith, K.P, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2018-04-23 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7180779 Å) | | Cite: | Insight into human Miro1/2 domain organization based on the structure of its N-terminal GTPase.
J.Struct.Biol., 212, 2020
|
|
6DUP
 
 | | CRYSTAL STRUCTURE OF PXR IN COMPLEX WITH COMPOUND 7 | | Descriptor: | (2S)-2-({[3'-(trifluoromethyl)[1,1'-biphenyl]-4-yl]oxy}methyl)-2,3-dihydro-7H-[1,3]oxazolo[3,2-a]pyrimidin-7-one, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Chen, X, Zhang, Y, Mclean, L.R. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Amelioration of PXR-mediated CYP3A4 induction by mGluR2 modulators.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
1L9S
 
 | |
