1VRA
 
 | |
3DCN
 
 | | Glomerella cingulata apo cutinase | | Descriptor: | Cutinase | | Authors: | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | Deposit date: | 2008-06-04 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
1VRD
 
 | |
7N89
 
 | |
7NH7
 
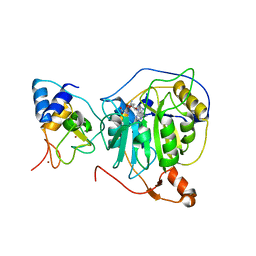 | |
5GZI
 
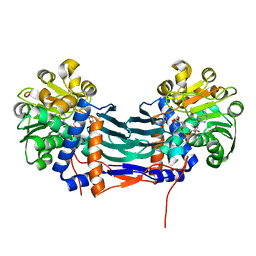 | | Cyclodeaminase_PA | | Descriptor: | (2S)-piperidine-2-carboxylic acid, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Ying, H, Chen, K. | | Deposit date: | 2016-09-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Cyclodeaminase_PA
To Be Published
|
|
5GQJ
 
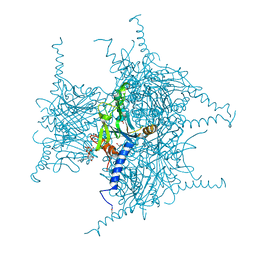 | | Crystal structure of Cypovirus Polyhedra mutant with deletion of Ser193 and Ala194 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
5GQM
 
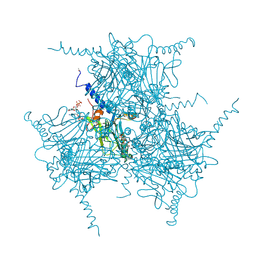 | | Crystal structure of in cellulo Wild Type Cypovirus Polyhedra | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
7RJ9
 
 | |
1VS0
 
 | | Crystal Structure of the Ligase Domain from M. tuberculosis LigD at 2.4A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative DNA ligase-like protein Rv0938/MT0965, ... | | Authors: | Akey, D, Martins, A, Aniukwu, J, Glickman, M.S, Shuman, S, Berger, J.M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-01-27 | | Release date: | 2006-02-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and Nonhomologous End-joining Function of the Ligase Component of Mycobacterium DNA Ligase D.
J.Biol.Chem., 281, 2006
|
|
3DEA
 
 | | Glomerella cingulata PETFP-cutinase complex | | Descriptor: | 1,1,1-trifluoro-3-[(2-phenylethyl)sulfanyl]propan-2-one, Cutinase | | Authors: | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | Deposit date: | 2008-06-09 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
3LH2
 
 | |
7RHA
 
 | | A new fluorescent protein darkmRuby at pH 5.0 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Huang, M, Ng, H.L, Zhang, S, Deng, M, Chu, J. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a new fluorescent protein darkmRuby at pH 5.0
To Be Published
|
|
7RKT
 
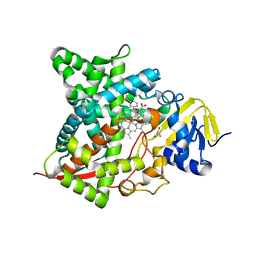 | |
7RHB
 
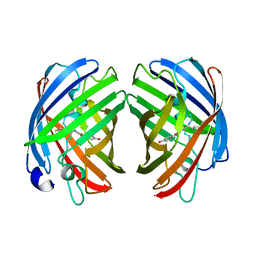 | |
111D
 
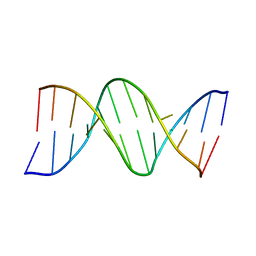 | |
7RHD
 
 | | darkmRuby M94T/F96Y mutant at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, darkmRuby M94T/F96Y mutant | | Authors: | Huang, M, Ng, H.L, Zhang, S, Deng, M, Chu, J. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Long-range Interaction Affects Brightness and pH Stability of a Dark Fluorescent Protein
To Be Published
|
|
1VER
 
 | | Structure of New Antigen Receptor variable domain from sharks | | Descriptor: | New Antigen Receptor variable domain | | Authors: | Streltsov, V.A. | | Deposit date: | 2004-04-05 | | Release date: | 2004-08-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural evidence for evolution of shark Ig new antigen receptor variable domain antibodies from a cell-surface receptor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1VGQ
 
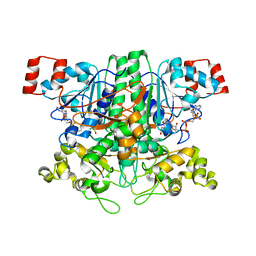 | | Formyl-CoA transferase mutant Asp169 to Ala | | Descriptor: | Formyl-coenzyme A transferase, OXIDIZED COENZYME A | | Authors: | Ricagno, S, Jonsson, S, Richards, N.G, Lindqvist, Y. | | Deposit date: | 2004-04-28 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes
J.Biol.Chem., 279, 2004
|
|
7RKW
 
 | |
7RHC
 
 | | A new fluorescent protein darkmRuby at pH 9.0 | | Descriptor: | 1,2-ETHANEDIOL, darkmRuby | | Authors: | Huang, M, Ng, H.L, Zhang, S, Deng, M, Chu, J. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Long-range Interaction Affects Brightness and pH Stability of a Dark Fluorescent Protein
To Be Published
|
|
7RKR
 
 | |
6DTU
 
 | | Maltotetraose bound T. maritima MalE1 | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE1 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
5GK7
 
 | | Structure of E.Coli fructose 1,6-bisphosphate aldolase bound to Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Fructose-bisphosphate aldolase class 2, ... | | Authors: | Tran, T.H, Huynh, K.H, Ho, T.H, Kang, L.W. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of E.Coli fructose 1,6-bisphosphate aldolase, Tris bound form
To Be Published
|
|
7RKD
 
 | |
