6RH4
 
 | | Human Carbonic Anhydrase II in complex with 4-Nitrobenzenesulfonamide. | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 4-nitrobenzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.948 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
8OE6
 
 | | Structure of hyperstable haloalkane dehalogenase variant DhaA231 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Structure of hyperstable haloalkane dehalogenase variant DhaA231 | | Authors: | Marek, M. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Advancing Enzyme's Stability and Catalytic Efficiency through Synergy of Force-Field Calculations, Evolutionary Analysis, and Machine Learning.
Acs Catalysis, 13, 2023
|
|
1R9D
 
 | | Glycerol bound form of the B12-independent glycerol dehydratase from Clostridium butyricum | | Descriptor: | GLYCEROL, glycerol dehydratase | | Authors: | Lanzilotta, W.N, O'Brien, J.R, Raynaud, C, Soucaille, P. | | Deposit date: | 2003-10-28 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insight into the mechanism of the B12-independent glycerol dehydratase from Clostridium butyricum: preliminary biochemical and structural characterization.
Biochemistry, 43, 2004
|
|
8OE2
 
 | | Structure of hyperstable haloalkane dehalogenase variant DhaA223 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Marek, M. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Advancing Enzyme's Stability and Catalytic Efficiency through Synergy of Force-Field Calculations, Evolutionary Analysis, and Machine Learning.
Acs Catalysis, 13, 2023
|
|
1R8W
 
 | |
8K5T
 
 | |
8K5S
 
 | |
1P18
 
 | | Hypoxanthine Phosphoribosyltransferase from Trypanosoma cruzi, K68R mutant, ternary substrates complex | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 7-HYDROXY-PYRAZOLO[4,3-D]PYRIMIDINE, MAGNESIUM ION, ... | | Authors: | Canyuk, B, Eakin, A.E, Craig III, S.P. | | Deposit date: | 2003-04-11 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interactions at the dimer interface influence the relative efficiencies for purine nucleotide synthesis and pyrophosphorolysis in a phosphoribosyltransferase
J.Mol.Biol., 335, 2004
|
|
5I4Q
 
 | | Contact-dependent inhibition system from Escherichia coli NC101 - ternary CdiA/CdiI/EF-Tu complex (domains 2 and 3) | | Descriptor: | CHLORIDE ION, Contact-dependent inhibitor A, Contact-dependent inhibitor I, ... | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2016-02-12 | | Release date: | 2017-06-28 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs.
Nucleic Acids Res., 45, 2017
|
|
1RF8
 
 | | Solution structure of the yeast translation initiation factor eIF4E in complex with m7GDP and eIF4GI residues 393 to 490 | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic initiation factor 4F subunit p150, Eukaryotic translation initiation factor 4E, ... | | Authors: | Gross, J.D, Moerke, N.J, von der Haar, T, Lugovskoy, A.A, Sachs, A.B, McCarthy, J.E.G, Wagner, G. | | Deposit date: | 2003-11-07 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-06 | | Method: | SOLUTION NMR | | Cite: | Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E.
Cell(Cambridge,Mass.), 115, 2003
|
|
3RSZ
 
 | | Maltodextran bound basal state conformation of yeast glycogen synthase isoform 2 | | Descriptor: | Glycogen [starch] synthase isoform 2, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Baskaran, S, Hurley, T.D. | | Deposit date: | 2011-05-02 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Multiple Glycogen-binding Sites in Eukaryotic Glycogen Synthase Are Required for High Catalytic Efficiency toward Glycogen.
J.Biol.Chem., 286, 2011
|
|
4GS7
 
 | | Structure of the Interleukin-15 quaternary complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ring, A.M, Ozkan, E, Feng, D, Garcia, K.C. | | Deposit date: | 2012-08-27 | | Release date: | 2012-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanistic and structural insight into the functional dichotomy between IL-2 and IL-15.
Nat.Immunol., 13, 2012
|
|
6MB5
 
 | | AAC-IIIb binary with NEOMYCIN | | Descriptor: | Aac(3)-IIIb protein, NEOMYCIN | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2018-08-29 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Encoding of Promiscuity in an Aminoglycoside Acetyltransferase.
J. Med. Chem., 61, 2018
|
|
1MXH
 
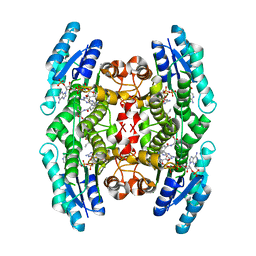 | | Crystal Structure of Substrate Complex of Putative Pteridine Reductase 2 (PTR2) from Trypanosoma cruzi | | Descriptor: | DIHYDROFOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 2 | | Authors: | Schormann, N, Pal, B, Senkovich, O, Carson, M, Howard, A, Smith, C, Delucas, L, Chattopadhyay, D. | | Deposit date: | 2002-10-02 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Trypanosoma cruzi pteridine reductase 2 in complex with a substrate and an inhibitor.
J.Struct.Biol., 152, 2005
|
|
1FJ0
 
 | |
6SD7
 
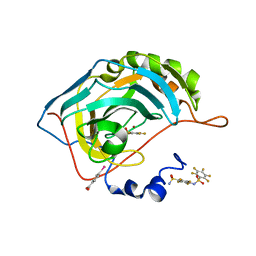 | | Human Carbonic Anhydrase II in complex with fluorinated benzenesulfonamide and its dimer | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 2,3,4,5,6-pentafluoro-N-(2,3,5,6-tetrafluoro-4-sulfamoylphenyl)benzenesulfonamide, 2,3,4,5,6-pentafluorobenzenesulfonamide, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-07-26 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
6SCH
 
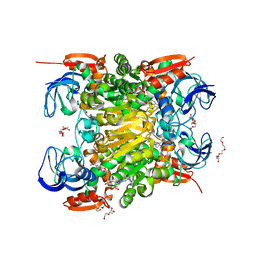 | | NADH-dependent variant of CBADH | | Descriptor: | MAGNESIUM ION, NADP-dependent isopropanol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Selles Vidal, L, Murray, J.W, Heap, J.T. | | Deposit date: | 2019-07-24 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Versatile selective evolutionary pressure using synthetic defect in universal metabolism.
Nat Commun, 12, 2021
|
|
4E3T
 
 | |
6K4K
 
 | |
6K4R
 
 | |
3TEH
 
 | | Crystal structure of Thermus thermophilus Phenylalanyl-tRNA synthetase complexed with L-dopa | | Descriptor: | 3,4-DIHYDROXYPHENYLALANINE, Phenylalanyl-tRNA synthetase alpha chain, Phenylalanyl-tRNA synthetase beta chain | | Authors: | Safro, M, Klipcan, L, Moor, N. | | Deposit date: | 2011-08-14 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8524 Å) | | Cite: | Bacterial and Eukaryotic Phenylalanyl-tRNA Synthetases Catalyze Misaminoacylation of tRNA(Phe) with 3,4-Dihydroxy-L-Phenylalanine.
Chem.Biol., 18, 2011
|
|
5TOV
 
 | |
6M7W
 
 | |
3GST
 
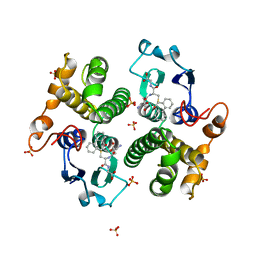 | | STRUCTURE OF THE XENOBIOTIC SUBSTRATE BINDING SITE OF A GLUTATHIONE S-TRANSFERASE AS REVEALED BY X-RAY CRYSTALLOGRAPHIC ANALYSIS OF PRODUCT COMPLEXES WITH THE DIASTEREOMERS OF 9-(S-GLUTATHIONYL)-10-HYDROXY-9, 10-DIHYDROPHENANTHRENE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Ji, X, Ammon, H.L, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the xenobiotic substrate binding site of a glutathione S-transferase as revealed by X-ray crystallographic analysis of product complexes with the diastereomers of 9-(S-glutathionyl)-10-hydroxy-9,10-dihydrophenanthrene.
Biochemistry, 33, 1994
|
|
5G1N
 
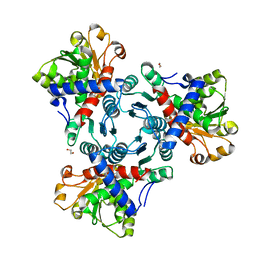 | | Aspartate transcarbamoylase domain of human CAD bound to PALA | | Descriptor: | 1,2-ETHANEDIOL, CAD PROTEIN, N-(PHOSPHONACETYL)-L-ASPARTIC ACID | | Authors: | Ruiz-Ramos, A, Grande-Garcia, A, Moreno-Morcillo, M.D, Ramon-Maiques, S. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Functional Characterization of Human Aspartate Transcarbamoylase, the Target of the Anti-Tumoral Drug Pala.
Structure, 24, 2016
|
|
