8B48
 
 | | Structure of Lentithecium fluviatile carbohydrate esterase from the CE15 family (LfCE15C) | | Descriptor: | Carbohydrate esterase family 15 protein, FORMIC ACID, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scholzen, K, Mazurkewich, S, Poulsen, J.C.N, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2022-09-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and functional investigation of a fungal member of carbohydrate esterase family 15 with potential specificity for rare xylans.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8WVT
 
 | | Crystal structure of Lsd18 in complex with ligands | | Descriptor: | (4R,5S)-3-((2R,3S,4S,E)-2,6-diethyl-9-((2R,3R)-2-ethyl-3-methyloxiran-2-yl)-3-hydroxy-4-methylnon-6-enoyl)-4-methyl-5-phenyloxazolidin-2-one, (4R,5S)-4-methyl-5-phenyl-3-((2R,3S,4S,6E,10E)-2,6,10-triethyl-3-hydroxy-4-methyldodeca-6,10-dienoyl)oxazolidin-2-one, CHLORIDE ION, ... | | Authors: | Deng, Y.M, Hu, Y.L, Chen, X. | | Deposit date: | 2023-10-24 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of Lsd18 in complex with ligands
To Be Published
|
|
5E29
 
 | | Crystal Structure of Deoxygenated Hemoglobin in Complex with an Allosteric Effector and Nitric Oxide | | Descriptor: | 2-(nitrooxy)ethyl 2-(4-{2-[(3,5-dimethylphenyl)amino]-2-oxoethyl}phenoxy)-2-methylpropanoate, 2-{4-[(3,5-DIMETHYLANILINO)-CARBONYL-METHYL]-PHENOXY}-2-METHYLPROPIONIC ACID, CACODYLATE ION, ... | | Authors: | Safo, M.K, Deshpande, T.M. | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design, Synthesis, and Investigation of Novel Nitric Oxide (NO)-Releasing Prodrugs as Drug Candidates for the Treatment of Ischemic Disorders: Insights into NO-Releasing Prodrug Biotransformation and Hemoglobin-NO Biochemistry.
Biochemistry, 54, 2015
|
|
5E83
 
 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN S (LIGANDED SICKLE CELL HEMOGLOBIN) COMPLEXED WITH GBT440, CO-CRYSTALLIZATION EXPERIMENT | | Descriptor: | 2-methyl-3-({2-[1-(propan-2-yl)-1H-pyrazol-5-yl]pyridin-3-yl}methoxy)phenol, CARBON MONOXIDE, GLYCEROL, ... | | Authors: | Patskovska, L, Patskovsky, Y, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-10-13 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | GBT440 increases haemoglobin oxygen affinity, reduces sickling and prolongs RBC half-life in a murine model of sickle cell disease.
Br.J.Haematol., 175, 2016
|
|
6ZK7
 
 | | Crystal Structure of human PYROXD1/FAD complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyridine nucleotide-disulfide oxidoreductase domain-containing protein 1 | | Authors: | Meinhart, A, Asanovic, I, Martinez, J, Clausen, T. | | Deposit date: | 2020-06-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The oxidoreductase PYROXD1 uses NAD(P) + as an antioxidant to sustain tRNA ligase activity in pre-tRNA splicing and unfolded protein response.
Mol.Cell, 81, 2021
|
|
2ZSE
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with AMPPCP and Pantothenate | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, GLYCEROL, ... | | Authors: | Chetnani, B, Das, S, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2008-09-05 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis pantothenate kinase: possible changes in location of ligands during enzyme action
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2ZSO
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 100 K: Laser on [450 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-17 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZS9
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with ADP and Pantothenate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, PANTOTHENOIC ACID, ... | | Authors: | Chetnani, B, Das, S, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2008-09-04 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mycobacterium tuberculosis pantothenate kinase: possible changes in location of ligands during enzyme action
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2ZSX
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 140 K: Laser on [600 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2AR5
 
 | |
2ZST
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 140 K: Laser on [450 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-17 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZT2
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 120 K: Laser on [600 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4ZBN
 
 | |
2ZS8
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) co-crystallized with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Pantothenate kinase | | Authors: | Chetnani, B, Das, S, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2008-09-03 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mycobacterium tuberculosis pantothenate kinase: possible changes in location of ligands during enzyme action
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2ZT4
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 120 K: Laser on [810 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4MRM
 
 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist phaclofen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-17 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
3OR1
 
 | | Crystal structure of dissimilatory sulfite reductase I (DsrI) | | Descriptor: | IRON/SULFUR CLUSTER, SIROHEME, SULFITE ION, ... | | Authors: | Hsieh, Y.C, Liu, M.Y, Wang, V.C.C, Chiang, Y.L, Liu, E.H, Wu, W.G, Chan, S.I, Chen, C.J. | | Deposit date: | 2010-09-06 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure insights into the enzyme catalysis from comparison of three forms of dissimilatory sulfite reductase from Desulfovibrio gigas
To be Published
|
|
5L8O
 
 | | crystal structure of human FABP6 in complex with cholate | | Descriptor: | CHOLIC ACID, DI(HYDROXYETHYL)ETHER, Gastrotropin | | Authors: | Hendrick, A, Mueller, I, Leonard, P.M, Davenport, R, Mitchell, P. | | Deposit date: | 2016-06-08 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Identification and Investigation of Novel Binding Fragments in the Fatty Acid Binding Protein 6 (FABP6).
J.Med.Chem., 59, 2016
|
|
7NZ5
 
 | | monomeric acetyl-CoA synthase with Zn at the proximal position of cluster A | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-[3,8,8,12,12-pentakis(2-hydroxy-2-oxoethyl)-2,7,11-tris(oxidanylidene)-1,4,6,9,10,13-hexaoxa-5$l^{6}-titanaspiro[4.4^{5}.4^{5}]tridecan-3-yl]ethanoic acid, CITRIC ACID, ... | | Authors: | Kreibich, J, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2021-03-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand binding at the Ni,Ni-[4Fe-4S] cluster of acetyl-CoA synthase
To Be Published
|
|
7O0D
 
 | |
4QI9
 
 | | Crystal structure of dihydrofolate reductase from Yersinia pestis complexed with methotrexate | | Descriptor: | Dihydrofolate reductase, METHOTREXATE | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structure of dihydrofolate reductase from Yersinia pestis complexed with methotrexate
To be Published
|
|
7NYP
 
 | | monomeric acetyl-CoA synthase in closed conformation | | Descriptor: | 2-[3,8,8,12,12-pentakis(2-hydroxy-2-oxoethyl)-2,7,11-tris(oxidanylidene)-1,4,6,9,10,13-hexaoxa-5$l^{6}-titanaspiro[4.4^{5}.4^{5}]tridecan-3-yl]ethanoic acid, CO-methylating acetyl-CoA synthase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kreibich, J, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2021-03-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand binding at the Ni,Ni-[4Fe-4S] cluster of acetyl-CoA synthase
To Be Published
|
|
7NYS
 
 | | monomeric acetyl-CoA synthase in closed conformation with carbon monoxide bound to the Ni proximal of cluster A | | Descriptor: | $l^{3}-oxidanylidynemethylnickel, 2-[3,8,8,12,12-pentakis(2-hydroxy-2-oxoethyl)-2,7,11-tris(oxidanylidene)-1,4,6,9,10,13-hexaoxa-5$l^{6}-titanaspiro[4.4^{5}.4^{5}]tridecan-3-yl]ethanoic acid, CHLORIDE ION, ... | | Authors: | Kreibich, J, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2021-03-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand binding at the Ni,Ni-[4Fe-4S] cluster of acetyl-CoA synthase
To Be Published
|
|
4MQF
 
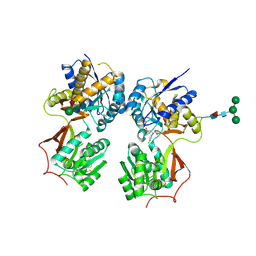 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist 2-hydroxysaclofen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-hydroxysaclofen, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-16 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
2NYK
 
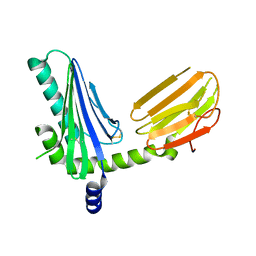 | | Crystal structure of m157 from mouse cytomegalovirus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, M157 | | Authors: | Garcia, K.C. | | Deposit date: | 2006-11-20 | | Release date: | 2007-05-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural elucidation of the m157 mouse cytomegalovirus ligand for Ly49 natural killer cell receptors.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
