6QCN
 
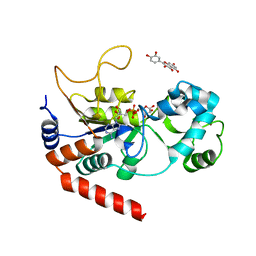 | | Human Sirt2 in complex with ADP-ribose and the inhibitor quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, NAD-dependent protein deacetylase sirtuin-2, ZINC ION, ... | | Authors: | Riemer, S, You, W, Steegborn, C. | | Deposit date: | 2018-12-29 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural basis for the activation and inhibition of Sirtuin 6 by quercetin and its derivatives.
Sci Rep, 9, 2019
|
|
1SMY
 
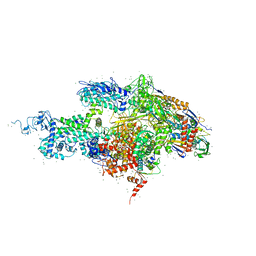 | | Structural basis for transcription regulation by alarmone ppGpp | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Patlan, V, Sekine, S, Vassylyeva, M.N, Hosaka, T, Ochi, K, Yokoyama, S, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-10 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for transcription regulation by alarmone ppGpp
Cell(Cambridge,Mass.), 117, 2004
|
|
1JCI
 
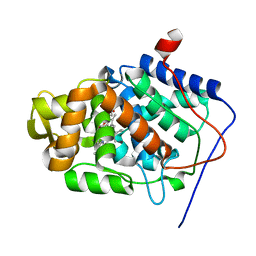 | | Stabilization of the Engineered Cation-binding Loop in Cytochrome c Peroxidase (CcP) | | Descriptor: | Cytochrome C Peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bhaskar, B, Bonagura, C.A, Li, H, Poulos, T.L. | | Deposit date: | 2001-06-09 | | Release date: | 2002-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cation-induced stabilization of the engineered cation-binding loop in cytochrome c peroxidase (CcP).
Biochemistry, 41, 2002
|
|
6QCT
 
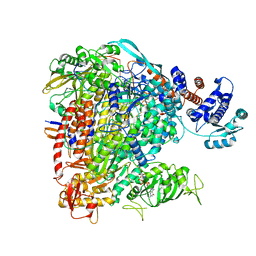 | | Influenza B polymerase elongation complex | | Descriptor: | 3 end, 5 end, MAGNESIUM ION, ... | | Authors: | Cusack, S, Kouba, T. | | Deposit date: | 2018-12-30 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural snapshots of actively transcribing influenza polymerase.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3MRW
 
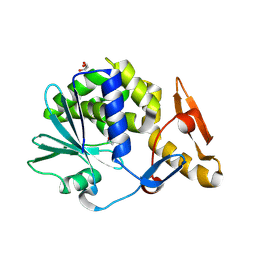 | | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina at 1.7 A resolution
To be Published
|
|
7STV
 
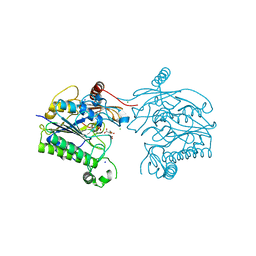 | | Crystal structure of sulfatase from Pedobacter yulinensis | | Descriptor: | CALCIUM ION, CHLORIDE ION, CITRIC ACID, ... | | Authors: | O'Malley, A, Schlachter, C.R, Grimes, L.L, Tomashek, J.J, Lee, A.L, Chruszcz, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis .
Molecules, 27, 2021
|
|
1SO9
 
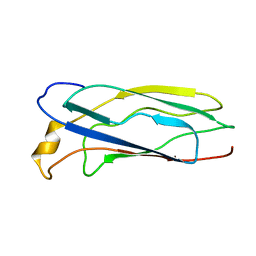 | | Solution Structure of apoCox11, 30 structures | | Descriptor: | Cytochrome C oxidase assembly protein ctaG | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
7SQ9
 
 | | Cryo-EM structure of mouse temsirolimus/PI(3,5)P2-bound TRPML1 channel at 2.11 Angstrom resolution | | Descriptor: | (1R,2R,4S)-4-{(2R)-2-[(3S,6R,7E,9R,10R,12R,14S,15E,17E,19E,21S,23S,26R,27R,30S,34aS)-9,27-dihydroxy-10,21-dimethoxy-6,8,12,14,20,26-hexamethyl-1,5,11,28,29-pentaoxo-1,4,5,6,9,10,11,12,13,14,21,22,23,24,25,26,27,28,29,31,32,33,34,34a-tetracosahydro-3H-23,27-epoxypyrido[2,1-c][1,4]oxazacyclohentriacontin-3-yl]propyl}-2-methoxycyclohexyl 3-hydroxy-2-(hydroxymethyl)-2-methylpropanoate, (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gan, N, Han, Y, Jiang, Y. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Structural mechanism of allosteric activation of TRPML1 by PI(3,5)P 2 and rapamycin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6QCZ
 
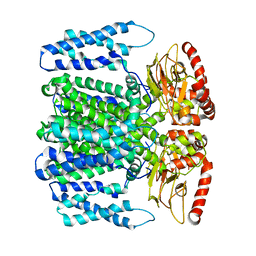 | | MloK1 model from single particle analysis of 2D crystals, class 3 (intermediate extended conformation) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Righetto, R, Biyani, N, Kowal, J, Chami, M, Stahlberg, H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM.
Nat Commun, 10, 2019
|
|
3MS7
 
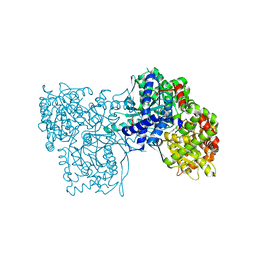 | |
1JCT
 
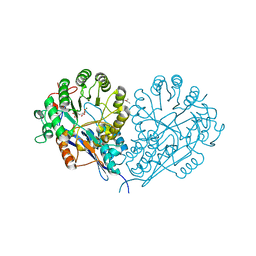 | | Glucarate Dehydratase, N341L mutant Orthorhombic Form | | Descriptor: | D-GLUCARATE, Glucarate Dehydratase, ISOPROPYL ALCOHOL, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2001-06-11 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in the active site of D-glucarate dehydratase from Escherichia coli.
Biochemistry, 40, 2001
|
|
1JBI
 
 | | NMR structure of the LCCL domain | | Descriptor: | cochlin | | Authors: | Liepinsh, E, Trexler, M, Kaikkonen, A, Weigelt, J, Banyai, L, Patthy, L, Otting, G. | | Deposit date: | 2001-06-05 | | Release date: | 2001-10-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the LCCL domain and implications for DFNA9 deafness disorder.
EMBO J., 20, 2001
|
|
7T7C
 
 | |
1SGM
 
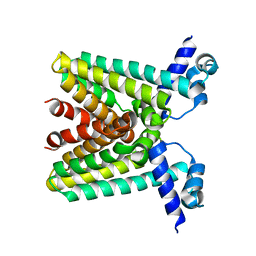 | |
6QD3
 
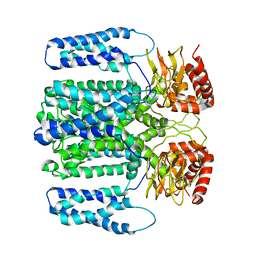 | | MloK1 model from single particle analysis of 2D crystals, class 7 (intermediate conformation) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Righetto, R, Biyani, N, Kowal, J, Chami, M, Stahlberg, H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM.
Nat Commun, 10, 2019
|
|
7TC0
 
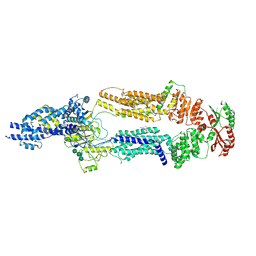 | | The structure of human ABCA1 in digitonin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATP-binding cassette, ... | | Authors: | Sun, Y, Li, X. | | Deposit date: | 2021-12-22 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cholesterol efflux mechanism revealed by structural analysis of human ABCA1 conformational states.
Nat Cardiovasc Res, 1, 2022
|
|
1JCG
 
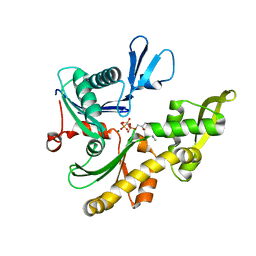 | | MREB FROM THERMOTOGA MARITIMA, AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ROD SHAPE-DETERMINING PROTEIN MREB | | Authors: | van den Ent, F, Amos, L.A, Lowe, J. | | Deposit date: | 2001-06-09 | | Release date: | 2001-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Prokaryotic origin of the actin cytoskeleton.
Nature, 413, 2001
|
|
6QCE
 
 | |
7STT
 
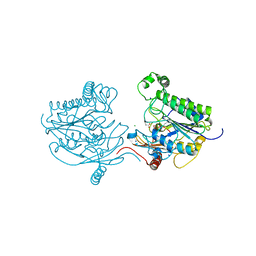 | | Crystal structure of sulfatase from Pedobacter yulinensis | | Descriptor: | CALCIUM ION, CHLORIDE ION, MALONATE ION, ... | | Authors: | O'Malley, A, Schlachter, C.R, Grimes, L.L, Tomashek, J.J, Lee, A.L, Chruszcz, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis .
Molecules, 27, 2021
|
|
1JCO
 
 | | Solution structure of the monomeric [Thr(B27)->Pro,Pro(B28)->Thr] insulin mutant (PT insulin) | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Keller, D, Clausen, R, Josefsen, K, Led, J.J. | | Deposit date: | 2001-06-11 | | Release date: | 2001-10-03 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Flexibility and bioactivity of insulin: an NMR investigation of the solution structure and folding of an unusually flexible human insulin mutant with increased biological activity.
Biochemistry, 40, 2001
|
|
6QCH
 
 | |
7TD0
 
 | | Lysophosphatidic acid receptor 1-Gi complex bound to LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
6QDI
 
 | | anti-sigma factor domain-containing protein from Clostridium clariflavum | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PA14 domain-containing protein | | Authors: | Voronov, M, Bayer, E.A, Livnah, O. | | Deposit date: | 2019-01-01 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Distinctive ligand-binding specificities of tandem PA14 biomass-sensory elements from Clostridium thermocellum and Clostridium clariflavum.
Proteins, 87, 2019
|
|
1JDD
 
 | | MUTANT (E219Q) MALTOTETRAOSE-FORMING EXO-AMYLASE COCRYSTALLIZED WITH MALTOTETRAOSE (CRYSTAL TYPE 2) | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
1JD9
 
 | |
