2W4W
 
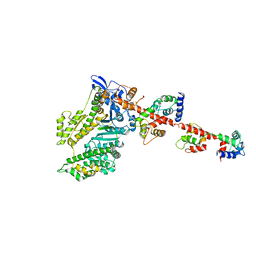 | | Isometrically contracting insect asynchronous flight muscle quick frozen after a quick stretch step | | Descriptor: | MYOSIN ESSENTIAL LIGHT CHAIN, STRIATED ADDUCTOR MUSCLE, MYOSIN HEAVY CHAIN, ... | | Authors: | Wu, S, Liu, J, Reedy, M.C, Tregear, R.T, Winkler, H, Franzini-Armstrong, C, Sasaki, H, Lucaveche, C, Goldman, Y.E, Reedy, M.K, Taylor, K.A. | | Deposit date: | 2008-12-02 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | Structural Changes in Isometrically Contracting Insect Flight Muscle Trapped Following a Mechanical Perturbation.
Plos One, 7, 2012
|
|
2W4T
 
 | | ISOMETRICALLY CONTRACTING INSECT ASYNCHRONOUS FLIGHT MUSCLE | | Descriptor: | MYOSIN ESSENTIAL LIGHT CHAIN, STRIATED ADDUCTOR MUSCLE, MYOSIN HEAVY CHAIN, ... | | Authors: | Wu, S, Liu, J, Reedy, M.C, Tregear, R.T, Winkler, H, Franzini-Armstrong, C, Sasaki, H, Lucaveche, C, Goldman, Y.E, Reedy, M.K, Taylor, K.A. | | Deposit date: | 2008-12-02 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | Electron Tomography of Cryofixed, Isometrically Contracting Insect Flight Muscle Reveals Novel Actin-Myosin Interactions
Plos One, 5, 2010
|
|
8DIT
 
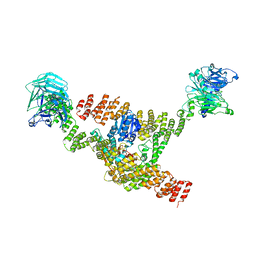 | | Cryo-EM structure of a HOPS core complex containing Vps33, Vps16, and Vps18 | | Descriptor: | Vacuolar protein sorting-associated protein 16, Vacuolar protein sorting-associated protein 18, Vacuolar protein sorting-associated protein 33 | | Authors: | Port, S.A, Farrell, P.D, Jeffrey, P.D, DiMaio, F, Hughson, F.M. | | Deposit date: | 2022-06-29 | | Release date: | 2022-08-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Cryo-EM structure of the HOPS core complex and its implication for SNARE assembly
To Be Published
|
|
4KMO
 
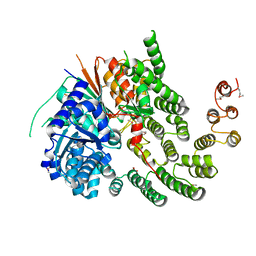 | | Crystal Structure of the Vps33-Vps16 HOPS subcomplex from Chaetomium thermophilum | | Descriptor: | Putative vacuolar protein sorting-associated protein, SULFATE ION, Small conjugating protein ligase-like protein | | Authors: | Baker, R.W, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2013-05-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Sec1/Munc18 (SM) Protein Vps33, Alone and Bound to the Homotypic Fusion and Vacuolar Protein Sorting (HOPS) Subunit Vps16*
Plos One, 8, 2013
|
|
1MOL
 
 | |
8JZ0
 
 | |
8JZ1
 
 | |
1FA3
 
 | |
2HDM
 
 | |
2JP1
 
 | |
2N54
 
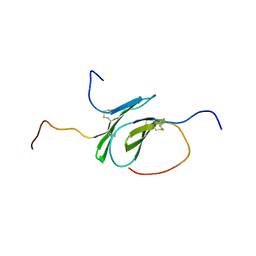 | | Solution structure of a disulfide stabilized XCL1 dimer | | Descriptor: | Lymphotactin | | Authors: | Tyler, R.C, Tuinstra, R.L, Peterson, F.F, Volkman, B.F. | | Deposit date: | 2015-07-07 | | Release date: | 2015-10-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Engineering Metamorphic Chemokine Lymphotactin/XCL1 into the GAG-Binding, HIV-Inhibitory Dimer Conformation.
Acs Chem.Biol., 10, 2015
|
|
2NYZ
 
 | |
7N2V
 
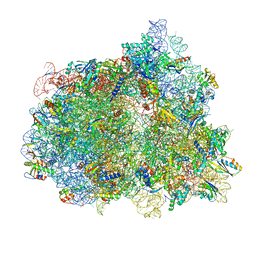 | | Elongating 70S ribosome complex in a spectinomycin-stalled intermediate state of translocation bound to EF-G in an active, GTP conformation (INT1) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-29 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
4C5G
 
 | |
4C5E
 
 | |
4C5H
 
 | |
7PIB
 
 | | 70S ribosome with EF-G, A/P- and P/E-site tRNAs in spectinomycin-treated Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-08-19 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
8CAI
 
 | | Streptomycin and Hygromycin B bound to the 30S body | | Descriptor: | 16S rRNA, 30S ribosomal protein S16, 30S ribosomal protein S2, ... | | Authors: | Paternoga, H, Crowe-McAuliffe, C, Novacek, J, Wilson, D.N. | | Deposit date: | 2023-01-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CA7
 
 | | Omadacycline and spectinomycin bound to the 30S ribosomal subunit head | | Descriptor: | 16S rRNA, 30S ribosomal protein S7, MAGNESIUM ION, ... | | Authors: | Paternoga, H, Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2023-01-24 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.06 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2W0T
 
 | |
7P7S
 
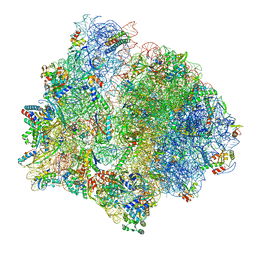 | | PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S ribosome, state II | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2021-07-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics.
Nat Commun, 13, 2022
|
|
7P7U
 
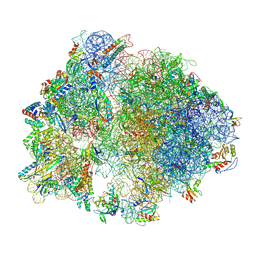 | | E. faecalis 70S ribosome with P-tRNA, state IV | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2021-07-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics.
Nat Commun, 13, 2022
|
|
7P7R
 
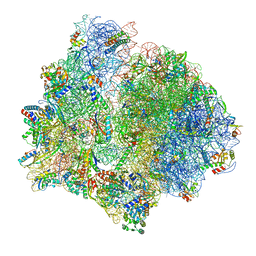 | | PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S ribosome, state I | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2021-07-20 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics.
Nat Commun, 13, 2022
|
|
7P7T
 
 | | PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S ribosome, state III | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2021-07-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics.
Nat Commun, 13, 2022
|
|
7P7Q
 
 | | E. faecalis 70S ribosome bound by PoxtA-EQ2, high-resolution combined volume | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2021-07-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics.
Nat Commun, 13, 2022
|
|
