3L2B
 
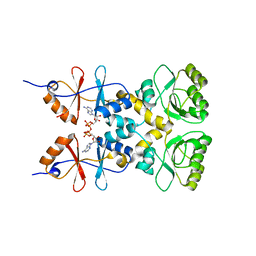 | | Crystal structure of the CBS and DRTGG domains of the regulatory region of Clostridium perfringens pyrophosphatase complexed with activator, diadenosine tetraphosphate | | Descriptor: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Probable manganase-dependent inorganic pyrophosphatase | | Authors: | Tuominen, H, Salminen, A, Oksanen, E, Jamsen, J, Heikkila, O, Lehtio, L, Magretova, N.N, Goldman, A, Baykov, A.A, Lahti, R. | | Deposit date: | 2009-12-15 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structures of the CBS and DRTGG Domains of the Regulatory Region of Clostridiumperfringens Pyrophosphatase Complexed with the Inhibitor, AMP, and Activator, Diadenosine Tetraphosphate.
J.Mol.Biol., 2010
|
|
3L31
 
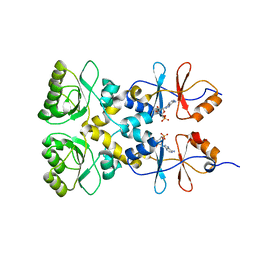 | | Crystal structure of the CBS and DRTGG domains of the regulatory region of Clostridium perfringens pyrophosphatase complexed with the inhibitor, AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Probable manganase-dependent inorganic pyrophosphatase | | Authors: | Tuominen, H, Salminen, A, Oksanen, E, Jamsen, J, Heikkila, O, Lehtio, L, Magretova, N.N, Goldman, A, Baykov, A.A, Lahti, R. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the CBS and DRTGG Domains of the Regulatory Region of Clostridiumperfringens Pyrophosphatase Complexed with the Inhibitor, AMP, and Activator, Diadenosine Tetraphosphate.
J.Mol.Biol., 2010
|
|
3LFR
 
 | |
3LFZ
 
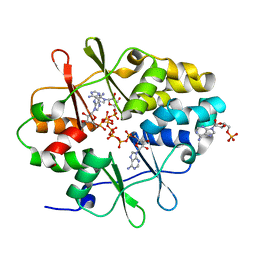 | | Crystal Structure of Protein MJ1225 from Methanocaldococcus jannaschii, a putative archaeal homolog of g-AMPK. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gomez-Garcia, I, Oyenarte, I, Kortazar, D, Martinez-Cruz, L.A. | | Deposit date: | 2010-01-19 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of protein MJ1225 from Methanocaldococcus jannaschii shows strong conservation of key structural features seen in the eukaryal gamma-AMPK.
J.Mol.Biol., 65, 2010
|
|
3LHH
 
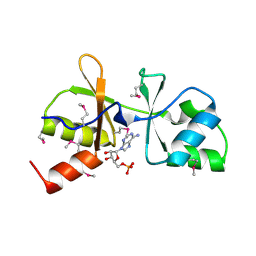 | | The crystal structure of CBS domain protein from Shewanella oneidensis MR-1. | | Descriptor: | ADENOSINE MONOPHOSPHATE, CBS domain protein | | Authors: | Tan, K, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of CBS domain protein from Shewanella oneidensis MR-1.
To be Published
|
|
3LQN
 
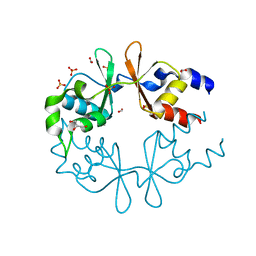 | | Crystal Structure of CBS Domain-containing Protein of Unknown Function from Bacillus anthracis str. Ames Ancestor | | Descriptor: | CBS domain protein, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Mulligan, R, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-09 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of CBS Domain-containing Protein of Unknown Function from Bacillus anthracis str. Ames Ancestor
To be Published
|
|
3LV9
 
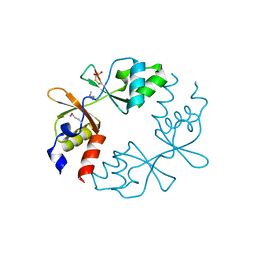 | |
3NQR
 
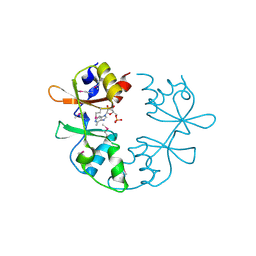 | | A putative CBS domain-containing protein from Salmonella typhimurium LT2 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Magnesium and cobalt efflux protein corC | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Cui, H, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-29 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A putative CBS domain-containing protein from Salmonella typhimurium LT2.
To be Published
|
|
3OCM
 
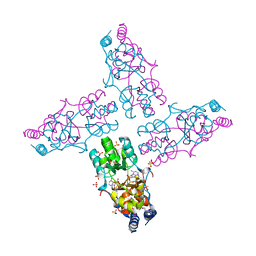 | | The crystal structure of a domain from a possible membrane protein of Bordetella parapertussis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative membrane protein, ... | | Authors: | Tan, K, Tesar, C, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-10 | | Release date: | 2010-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The crystal structure of a domain from a possible membrane protein of Bordetella parapertussis
To be Published
|
|
3OCO
 
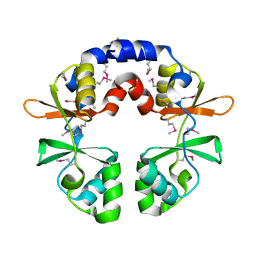 | |
3OI8
 
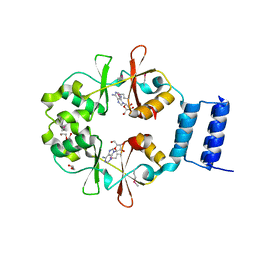 | | The crystal structure of functionally unknown conserved protein domain from Neisseria meningitidis MC58 | | Descriptor: | ADENOSINE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zhang, R, Tan, K, Li, H, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-18 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | The crystal structure of functionally unknown conserved protein domain from Neisseria meningitidis MC58
To be Published
|
|
3PC2
 
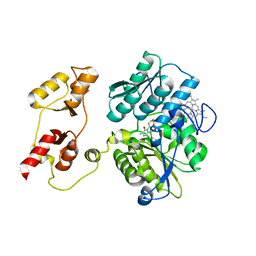 | |
3PC3
 
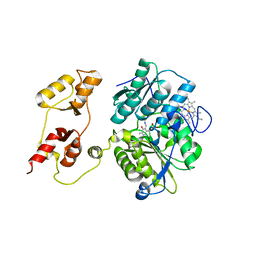 | |
3PC4
 
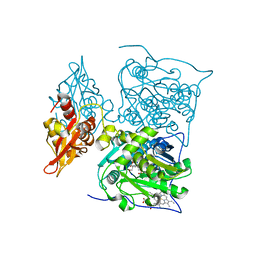 | |
3SL7
 
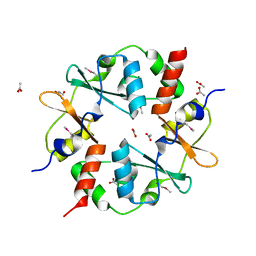 | | Crystal structure of CBS-pair protein, CBSX2 from Arabidopsis thaliana | | Descriptor: | ACETATE ION, CBS domain-containing protein CBSX2, GLYCEROL | | Authors: | Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2011-06-24 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Single cystathionine beta-synthase domain-containing proteins modulate development by regulating the thioredoxin system in Arabidopsis
Plant Cell, 23, 2011
|
|
3T4N
 
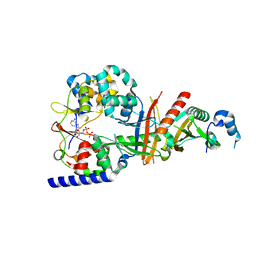 | | Structure of the regulatory fragment of Saccharomyces cerevisiae AMPK in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmdit, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-07-26 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
3TDH
 
 | | Structure of the regulatory fragment of sccharomyces cerevisiae AMPK in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmidt, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-08-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
3TE5
 
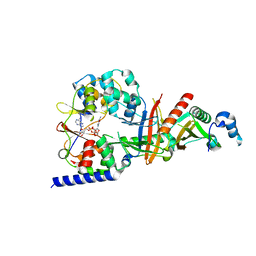 | | structure of the regulatory fragment of sacchromyces cerevisiae ampk in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmidt, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-08-12 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
3TSB
 
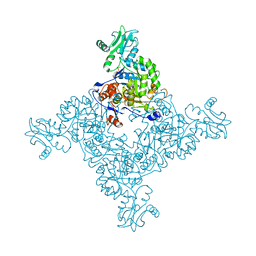 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames | | Descriptor: | Inosine-5'-monophosphate dehydrogenase, PHOSPHATE ION | | Authors: | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-12 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
3TSD
 
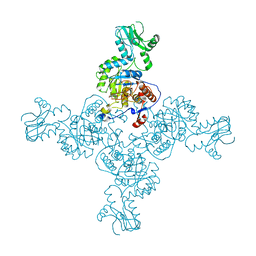 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames complexed with XMP | | Descriptor: | D(-)-TARTARIC ACID, Inosine-5'-monophosphate dehydrogenase, SULFATE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
3USB
 
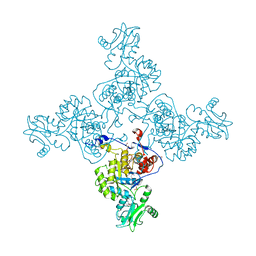 | | Crystal Structure of Bacillus anthracis Inosine Monophosphate Dehydrogenase in the complex with IMP | | Descriptor: | CHLORIDE ION, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Zhang, R, Wu, R, Gu, M, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
3ZFH
 
 | | Crystal structure of Pseudomonas aeruginosa inosine 5'-monophosphate dehydrogenase | | Descriptor: | CHLORIDE ION, INOSINE 5'-MONOPHOSPHATE DEHYDROGENASE | | Authors: | Rao, V.A, Shepherd, S.M, Owen, R, Hunter, W.N. | | Deposit date: | 2012-12-11 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Pseudomonas Aeruginosa Inosine 5'-Monophosphate Dehydrogenase
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4AF0
 
 | | Crystal structure of cryptococcal inosine monophosphate dehydrogenase | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID, MYCOPHENOLIC ACID, ... | | Authors: | Valkov, E, Stamp, A, Morrow, C.A, Kobe, B, Fraser, J.A. | | Deposit date: | 2012-01-15 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | De Novo GTP Biosynthesis is Critical for Virulence of the Fungal Pathogen Cryptococcus Neoformans
Plos Pathog., 8, 2012
|
|
4AVF
 
 | | Crystal structure of Pseudomonas aeruginosa inosine 5'-monophosphate dehydrogenase | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE | | Authors: | McMahon, S.A, Moynie, L, Liu, H, Duthie, F, Naismith, J.H. | | Deposit date: | 2012-05-25 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4CFE
 
 | | Structure of full length human AMPK in complex with a small molecule activator, a benzimidazole derivative (991) | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Carmena, D, Bright, N.J, Haire, L.F, Underwood, E, Patel, B.R, Heath, R.B, Walker, P.A, Hallen, S, Giordanetto, F, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.023 Å) | | Cite: | Structural Basis of Ampk Regulation by Small Molecule Activators.
Nat.Commun., 4, 2013
|
|
