5Z1Z
 
 | | The apo-structure of D-lactate dehydrogenase from Escherichia coli | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2017-12-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria.
Biochemistry, 57, 2018
|
|
3K5P
 
 | |
5DN9
 
 | | Crystal structure of Candida boidinii formate dehydrogenase complexed with NAD+ and azide | | Descriptor: | AZIDE ION, CHLORIDE ION, FDH, ... | | Authors: | Guo, Q, Gakhar, L, Wichersham, K, Francis, K, Vardi-Kilshtain, A, Major, D.T, Cheatum, C.M, Kohen, A. | | Deposit date: | 2015-09-09 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Kinetic Studies of Formate Dehydrogenase from Candida boidinii.
Biochemistry, 55, 2016
|
|
2YQ4
 
 | |
3KB6
 
 | | Crystal structure of D-Lactate dehydrogenase from aquifex aeolicus complexed with NAD and Lactic acid | | Descriptor: | D-lactate dehydrogenase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Ellis, M.J, Bessho, Y, Kuramitsu, S, Yokoyama, S, Hasnain, S.S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-20 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of D-lactate dehydrogenase from Aquifex aeolicus complexed with NAD(+) and lactic acid (or pyruvate).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
6ABI
 
 | | The apo-structure of D-lactate dehydrogenase from Fusobacterium nucleatum | | Descriptor: | D-lactate dehydrogenase, GLYCEROL, SULFATE ION | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2018-07-21 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria
Biochemistry, 57, 2018
|
|
3BAZ
 
 | | Structure of hydroxyphenylpyruvate reductase from coleus blumei in complex with NADP+ | | Descriptor: | Hydroxyphenylpyruvate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Janiak, V, Klebe, G, Petersen, M, Heine, A. | | Deposit date: | 2007-11-09 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and substrate docking of a hydroxy(phenyl)pyruvate reductase from the higher plant Coleus blumei Benth
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3BA1
 
 | | Structure of hydroxyphenylpyruvate reductase from coleus blumei | | Descriptor: | Hydroxyphenylpyruvate reductase | | Authors: | Janiak, V, Klebe, G, Petersen, M, Heine, A. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure and substrate docking of a hydroxy(phenyl)pyruvate reductase from the higher plant Coleus blumei Benth.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1J49
 
 | | INSIGHTS INTO DOMAIN CLOSURE, SUBSTRATE SPECIFICITY AND CATALYSIS OF D-LACTATE DEHYDROGENASE FROM LACTOBACILLUS BULGARICUS | | Descriptor: | D-LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Razeto, A, Kochhar, S, Hottinger, H, Dauter, M, Wilson, K.S, Lamzin, V.S. | | Deposit date: | 2001-08-14 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Domain closure, substrate specificity and catalysis of D-lactate dehydrogenase from Lactobacillus bulgaricus.
J.Mol.Biol., 318, 2002
|
|
4XKJ
 
 | |
4NU6
 
 | | Crystal Structure of PTDH R301K | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Phosphonate dehydrogenase, SULFATE ION | | Authors: | Nair, S.K, Chekan, J.R. | | Deposit date: | 2013-12-03 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Chemical rescue and inhibition studies to determine the role of arg301 in phosphite dehydrogenase.
Plos One, 9, 2014
|
|
4XYG
 
 | |
4XYB
 
 | | GRANULICELLA M. FORMATE DEHYDROGENASE (FDH) IN COMPLEX WITH NADP(+) AND NaN3 | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, Formate dehydrogenase, ... | | Authors: | Cendron, L, Fogal, S, Beneventi, E, Bergantino, E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural basis for double cofactor specificity in a new formate dehydrogenase from the acidobacterium Granulicella mallensis MP5ACTX8.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4XYE
 
 | | GRANULICELLA M. FORMATE DEHYDROGENASE (FDH) IN COMPLEX WITH NAD(+) | | Descriptor: | Formate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Cendron, L, Fogal, S, Beneventi, E, Bergantino, E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for double cofactor specificity in a new formate dehydrogenase from the acidobacterium Granulicella mallensis MP5ACTX8.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4NJO
 
 | |
4NU5
 
 | | Crystal Structure of PTDH R301A | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Phosphonate dehydrogenase | | Authors: | Nair, S.K, Chekan, J.R. | | Deposit date: | 2013-12-03 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemical rescue and inhibition studies to determine the role of arg301 in phosphite dehydrogenase.
Plos One, 9, 2014
|
|
4ZGS
 
 | | Identification of the pyruvate reductase of Chlamydomonas reinhardtii | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative D-lactate dehydrogenase | | Authors: | Burgess, S.J, Hussein, T, Yeoman, J.A, Iamshanova, O, Boehm, M, Bundy, J, Bialek, W, Murray, J.W, Nixon, P.J. | | Deposit date: | 2015-04-23 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | Identification of the Elusive Pyruvate Reductase of Chlamydomonas reinhardtii Chloroplasts.
Plant Cell.Physiol., 57, 2016
|
|
4NFY
 
 | |
4NJM
 
 | |
6ABJ
 
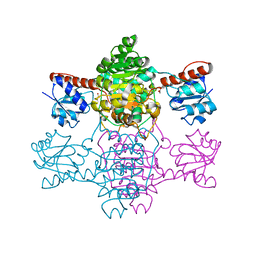 | | The apo-structure of D-lactate dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, D-lactate dehydrogenase (Fermentative) | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2018-07-21 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria
Biochemistry, 57, 2018
|
|
3DC2
 
 | |
3DDN
 
 | |
5TX7
 
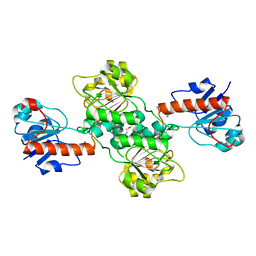 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Desulfovibrio vulgaris | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase family protein, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Czub, M.P, Shabalin, I.G, Gasiorowska, O.A, Handing, K.B, Kutner, J, Cymborowski, M.T, Hennig, P.M, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-11-15 | | Release date: | 2016-12-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Desulfovibrio vulgaris
to be published
|
|
3FN4
 
 | | Apo-form of NAD-dependent formate dehydrogenase from bacterium Moraxella sp.C-1 in closed conformation | | Descriptor: | GLYCEROL, NAD-dependent formate dehydrogenase, SULFATE ION | | Authors: | Shabalin, I.G, Polyakov, K.M, Filippova, E.V, Dorovatovskiy, P.V, Tikhonova, T.V, Sadykhov, E.G, Tishkov, V.I, Popov, V.O. | | Deposit date: | 2008-12-23 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of the apo and holo forms of formate dehydrogenase from the bacterium Moraxella sp. C-1: towards understanding the mechanism of the closure of the interdomain cleft
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6CDF
 
 | | Human CtBP1 (28-378) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C-terminal-binding protein 1, CALCIUM ION, ... | | Authors: | Royer, W.E, Bellesis, A.G. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Assembly of human C-terminal binding protein (CtBP) into tetramers.
J. Biol. Chem., 293, 2018
|
|
