7QMX
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 7) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QN0
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 10) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QN2
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 12) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6UJ0
 
 | | Unbound BACE2 mutant structure | | Descriptor: | Beta-secretase 2, unidentified polypeptide | | Authors: | Yen, Y.C, Ghosh, A.K, Mesecar, A.D. | | Deposit date: | 2019-10-01 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Structure-Based Discovery Platform for BACE2 and the Development of Selective BACE Inhibitors.
Acs Chem Neurosci, 12, 2021
|
|
6UWP
 
 | | BACE-1 in complex with compound #32 | | Descriptor: | (1R,2R)-2-[(4aR,7aR)-2-amino-6-(pyrimidin-2-yl)-4a,5,6,7-tetrahydropyrrolo[3,4-d][1,3]thiazin-7a(4H)-yl]-N-{[(1R,2R)-2-methylcyclopropyl]methyl}cyclopropane-1-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6UVP
 
 | | BACE-1 in complex with compound #3 | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{(1R,2R)-2-[(4aS,7aR)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]cyclopropyl}-5-fluoropyridine-2-carboxamide, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-04 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6UWV
 
 | | BACE-1 in complex with compound #34 | | Descriptor: | (4aR,7aR)-7a-[(1R,2R)-2-(2-{[(1R,2R)-2-methylcyclopropyl]methoxy}propan-2-yl)cyclopropyl]-6-(pyrimidin-2-yl)-4,4a,5,6,7,7a-hexahydropyrrolo[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Stout, S.L. | | Deposit date: | 2019-11-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6UVY
 
 | | BACE-1 in complex with compound #18 | | Descriptor: | (1R,2R)-2-[(4aS,7aR)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]-N-{[(1R,2R)-2-methylcyclopropyl]methyl}cyclopropane-1-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-04 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6UVV
 
 | | BACE-1 in complex with compound #17 | | Descriptor: | (1R,2R)-2-[(4aS,7aR)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]-N-butylcyclopropane-1-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-04 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6UJ1
 
 | | BACE2 mutant in complex with a macrocyclic compound | | Descriptor: | (3S)-3-hydroxy-N-(2-methylpropyl)-N~2~-{[(4S)-17-[(methylsulfonyl)(propyl)amino]-2-oxo-3-azatricyclo[13.3.1.1~6,10~]icosa-1(19),6(20),7,9,15,17-hexaen-4-yl]methyl}-L-norleucinamide, Beta-secretase 2 | | Authors: | Yen, Y.C, Ghosh, A.K, Mesecar, A.D. | | Deposit date: | 2019-10-02 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | A Structure-Based Discovery Platform for BACE2 and the Development of Selective BACE Inhibitors.
Acs Chem Neurosci, 12, 2021
|
|
6JSE
 
 | | Crystal Structure of BACE1 in complex with N-(3-((4S,5R)-2-amino-4-methyl-5-phenyl-5,6-dihydro-4H-1,3-thiazin-4-yl)-4-fluorophenyl)-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
6JSF
 
 | | Crystal Structure of BACE1 in complex with N-(3-((4S,5S)-2-amino-4-methyl-5-phenyl-5,6-dihydro-4H-1,3-thiazin-4-yl)-4-fluorophenyl)-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
6JT3
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4R,5R,6R)-2-amino-5-fluoro-4,6-dimethyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tadano, G, Komano, K, Yoshida, S, Suzuki, S, Nakahara, K, Fuchino, K, Fujimoto, K, Matsuoka, E, Yamamoto, T, Asada, N, Ito, H, Sakaguchi, G, Kanegawa, N, Kido, Y, Ando, S, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Bergh, A.V.D, Austin, N, Gijsen, H.J.M, Yamano, Y, Iso, Y, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of an Extremely Potent Thiazine-Based beta-Secretase Inhibitor with Reduced Cardiovascular and Liver Toxicity at a Low Projected Human Dose.
J.Med.Chem., 62, 2019
|
|
6JSG
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S)-2-amino-4-methyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-chloropyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
6JSZ
 
 | | BACE2 xaperone complex with N-{3-[(5R)-3-amino-5-methyl-9,9-dioxo-2,9lambda6-dithia-4-azaspiro[5.5]undec-3-en-5-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 2, CHLORIDE ION, N-[3-[(5R)-3-azanyl-5-methyl-9,9-bis(oxidanylidene)-2,9$l^{6}-dithia-4-azaspiro[5.5]undec-3-en-5-yl]-4-fluoranyl-phenyl]-5-(fluoranylmethoxy)pyrazine-2-carboxamide, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
6JT4
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S,6S)-2-amino-4-methyl-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Anan, K, Iso, Y, Oguma, T, Nakahara, K, Suzuki, S, Yamamoto, T, Matsuoka, E, Ito, H, Sakaguchi, G, Ando, S, Morimoto, K, Kanegawa, N, Kido, Y, Kawachi, T, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Austin, N, Bergh, A.V.D, Verboven, P, Bischoff, F, Gijsen, H.J.M, Yamano, Y, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trifluoromethyl Dihydrothiazine-Based beta-Secretase (BACE1) Inhibitors with Robust Central beta-Amyloid Reduction and Minimal Covalent Binding Burden.
Chemmedchem, 14, 2019
|
|
6WNY
 
 | | Crystal structure of BACE1 in complex with (Z)-fluoro-olefin containing compound 15 | | Descriptor: | 6-[(Z)-2-{3-[(1S,5S,6S)-3-amino-5-methyl-1-(morpholine-4-carbonyl)-2-thia-4-azabicyclo[4.1.0]hept-3-en-5-yl]-4-fluorophenyl}-1-fluoroethenyl]pyridine-3-carbonitrile, Beta-secretase 1, IODIDE ION | | Authors: | Whittington, D.A. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The development of a structurally distinct series of BACE1 inhibitors via the (Z)-fluoro-olefin amide bioisosteric replacement.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6JSN
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(5R)-3-amino-5-methyl-9,9-dioxo-2,9lambda6-dithia-4-azaspiro[5.5]undec-3-en-5-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
8CIK
 
 | | Altai wapiti (Cervus elaphus sibiricus) chymosin at 2.2 A resolution | | Descriptor: | Chymosin, GLYCEROL | | Authors: | Diusenova, S.E, Shevtsov, M.B, Borshchevskiy, V.I, Belenkaya, S.V, Kolybalov, D.S, Arkhipov, S.G, Volosnikova, E.A, Elchaninov, V.V, Shcherbakov, D.N. | | Deposit date: | 2023-02-09 | | Release date: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Altai wapiti (Cervus elaphus sibiricus) chymosin at 2.2 A resolution
To Be Published
|
|
8C6P
 
 | | Fragment screening hit I bound to endothiapepsin | | Descriptor: | 4-[(2-azanyl-4-methyl-1,3-thiazol-5-yl)methyl]benzenecarbonitrile, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C6Q
 
 | | Fragment screening hit II bound to endothiapepsin | | Descriptor: | 3-(4-methylimidazol-1-yl)-5-(trifluoromethyl)aniline, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C71
 
 | | Pyrrolidine fragment 5b bound to endothiapepsin | | Descriptor: | (3~{R},4~{R})-4-(3,4-dihydro-1~{H}-isoquinolin-2-yl)pyrrolidin-3-ol, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C72
 
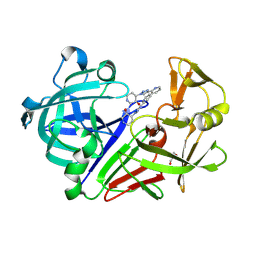 | | Pyrrolidine fragment 10b bound to endothiapepsin | | Descriptor: | (3~{S},4~{S})-4-(4-pyridin-2-yl-1,2,3-triazol-1-yl)piperidin-3-ol, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C70
 
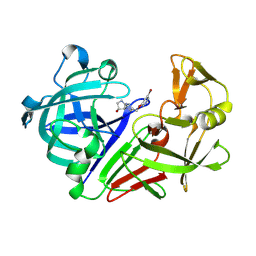 | | Pyrrolidine fragment 1 bound to endothiapepsin | | Descriptor: | (3~{R},4~{R})-4-[4-[(5-bromanylpyridin-3-yl)oxymethyl]-1,2,3-triazol-1-yl]pyrrolidin-3-ol, Endothiapepsin | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C74
 
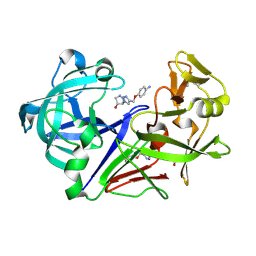 | | Pyrrolidine fragment 10d bound to endothiapepsin | | Descriptor: | (3~{R},4~{R})-4-[4-[(4-azanylphenoxy)methyl]-1,2,3-triazol-1-yl]pyrrolidin-3-ol, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
