1EXC
 
 | | CRYSTAL STRUCTURE OF B. SUBTILIS MAF PROTEIN COMPLEXED WITH D-(UTP) | | Descriptor: | DEOXYURIDINE-5'-TRIPHOSPHATE, PROTEIN MAF, SODIUM ION | | Authors: | Minasov, G, Teplova, M, Stewart, G.C, Koonin, E.V, Anderson, W.F, Egli, M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-05-02 | | Release date: | 2000-06-14 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Functional implications from crystal structures of the conserved Bacillus subtilis protein Maf with and without dUTP.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1F3E
 
 | | A NEW TARGET FOR SHIGELLOSIS: RATIONAL DESIGN AND CRYSTALLOGRAPHIC STUDIES OF INHIBITORS OF TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | 3,5-DIAMINOPHTHALHYDRAZIDE, QUEUINE TRNA-RIBOSYLTRANSFERASE, ZINC ION | | Authors: | Graedler, U, Gerber, H.-D, Goodenough-Lashua, D.M, Garcia, G.A.G, Ficner, R, Reuter, K, Stubbs, M.T, Klebe, G. | | Deposit date: | 2000-06-02 | | Release date: | 2000-06-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A new target for shigellosis: rational design and crystallographic studies of inhibitors of tRNA-guanine transglycosylase.
J.Mol.Biol., 306, 2001
|
|
1C8P
 
 | | NMR STRUCTURE OF THE LIGAND BINDING DOMAIN OF THE COMMON BETA-CHAIN IN THE GM-CSF, IL-3 AND IL-5 RECEPTORS | | Descriptor: | CYTOKINE RECEPTOR COMMON BETA CHAIN | | Authors: | Mulhern, T.D, D'Andrea, R.J, Gaunt, C, Vandeleur, L, Vadas, M.A, Lopez, A.F, Booker, G.W, Bagley, C.J. | | Deposit date: | 1999-10-05 | | Release date: | 2000-06-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the cytokine-binding domain of the common beta-chain of the receptors for granulocyte-macrophage colony-stimulating factor, interleukin-3 and interleukin-5.
J.Mol.Biol., 297, 2000
|
|
1QIQ
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ACmC Fe COMPLEX) | | Descriptor: | FE (III) ION, ISOPENICILLIN N SYNTHASE, N-[N-[2-AMINO-6-OXO-HEXANOIC ACID-6-YL]CYSTEINYL]-S-METHYLCYSTEINE, ... | | Authors: | Rutledge, P.J, Clifton, I.J, Burzlaff, N.I, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | Deposit date: | 1999-06-15 | | Release date: | 2000-06-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction.
Nature, 401, 1999
|
|
1D1F
 
 | |
1D0W
 
 | |
1D1E
 
 | |
1EJF
 
 | | CRYSTAL STRUCTURE OF THE HUMAN CO-CHAPERONE P23 | | Descriptor: | Prostaglandin E synthase 3, SULFATE ION | | Authors: | Weaver, A.J, Sullivan, W.P, Felts, S.J, Owen, B.A.L, Toft, D.O. | | Deposit date: | 2000-03-02 | | Release date: | 2000-06-19 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure and activity of human p23, a heat shock protein 90 co-chaperone.
J.Biol.Chem., 275, 2000
|
|
1EFW
 
 | | Crystal structure of aspartyl-tRNA synthetase from Thermus thermophilus complexed to tRNAasp from Escherichia coli | | Descriptor: | ASPARTYL-TRNA, ASPARTYL-TRNA SYNTHETASE | | Authors: | Briand, C, Poterszman, A, Eiler, S, Webster, G, Thierry, J.-C, Moras, D. | | Deposit date: | 2000-02-10 | | Release date: | 2000-06-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | An intermediate step in the recognition of tRNA(Asp) by aspartyl-tRNA synthetase.
J.Mol.Biol., 299, 2000
|
|
1QNN
 
 | |
1EWF
 
 | | THE 1.7 ANGSTROM CRYSTAL STRUCTURE OF BPI | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERICIDAL/PERMEABILITY-INCREASING PROTEIN | | Authors: | Kleiger, G, Beamer, L.J, Grothe, R, Mallick, P, Eisenberg, D. | | Deposit date: | 2000-04-25 | | Release date: | 2000-06-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7 A crystal structure of BPI: a study of how two dissimilar amino acid sequences can adopt the same fold.
J.Mol.Biol., 299, 2000
|
|
1EIK
 
 | | Solution Structure of RNA Polymerase Subunit RPB5 from Methanobacterium Thermoautotrophicum | | Descriptor: | RNA POLYMERASE SUBUNIT RPB5 | | Authors: | Yee, A, Booth, V, Dharamsi, A, Engel, A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-25 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA polymerase subunit RPB5 from Methanobacterium thermoautotrophicum.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EIS
 
 | | UDA UNCOMPLEXED FORM. CRYSTAL STRUCTURE OF URTICA DIOICA AGGLUTININ, A SUPERANTIGEN PRESENTED BY MHC MOLECULES OF CLASS I AND CLASS II | | Descriptor: | PROTEIN (AGGLUTININ ISOLECTIN VI/AGGLUTININ ISOLECTIN V) | | Authors: | Saul, F.A, Rovira, P, Boulot, G, Van Damme, E.J.M, Peumans, W.J, Truffa-Bachi, P, Bentley, G.A. | | Deposit date: | 2000-02-28 | | Release date: | 2000-06-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of Urtica dioica agglutinin, a superantigen presented by MHC molecules of class I and class II.
Structure Fold.Des., 8, 2000
|
|
1ENM
 
 | | UDA TRISACCHARIDE COMPLEX. CRYSTAL STRUCTURE OF URTICA DIOICA AGGLUTININ, A SUPERANTIGEN PRESENTED BY MHC MOLECULES OF CLASS I AND CLASS II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AGGLUTININ ISOLECTIN I/AGGLUTININ ISOLECTIN V/ AGGLUTININ ISOLECTIN VI | | Authors: | Saul, F.A, Rovira, P, Boulot, G, Van Damme, E.J.M, Peumans, W.J, Truffa-Bachi, P, Bentley, G.A. | | Deposit date: | 2000-03-21 | | Release date: | 2000-06-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Urtica dioica agglutinin, a superantigen presented by MHC molecules of class I and class II.
Structure Fold.Des., 8, 2000
|
|
1F0W
 
 | | CRYSTAL STRUCTURE OF ORTHORHOMBIC LYSOZYME GROWN AT PH 6.5 | | Descriptor: | LYSOZYME | | Authors: | Biswal, B.K, Sukumar, N, Vijayan, M. | | Deposit date: | 2000-05-17 | | Release date: | 2000-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hydration, mobility and accessibility of lysozyme: structures of a pH 6.5 orthorhombic form and its low-humidity variant and a comparative study involving 20 crystallographically independent molecules.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1EN2
 
 | | UDA TETRASACCHARIDE COMPLEX. CRYSTAL STRUCTURE OF URTICA DIOICA AGGLUTININ, A SUPERANTIGEN PRESENTED BY MHC MOLECULES OF CLASS I AND CLASS II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AGGLUTININ ISOLECTIN I/AGGLUTININ ISOLECTIN V/ AGGLUTININ ISOLECTIN VI | | Authors: | Saul, F.A, Rovira, P, Boulot, G, Van Damme, E.J.M, Peumans, W.J, Truffa-Bachi, P, Bentley, G.A. | | Deposit date: | 2000-03-20 | | Release date: | 2000-06-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Urtica dioica agglutinin, a superantigen presented by MHC molecules of class I and class II.
Structure Fold.Des., 8, 2000
|
|
1F03
 
 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
1F04
 
 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
1F10
 
 | |
1F4J
 
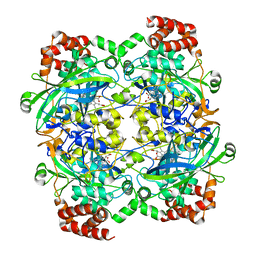 | | STRUCTURE OF TETRAGONAL CRYSTALS OF HUMAN ERYTHROCYTE CATALASE | | Descriptor: | CATALASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Safo, M.K, Musayev, F.N, Wu, S.H, Abraham, D.J, Ko, T.P. | | Deposit date: | 2000-06-07 | | Release date: | 2000-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of tetragonal crystals of human erythrocyte catalase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1F15
 
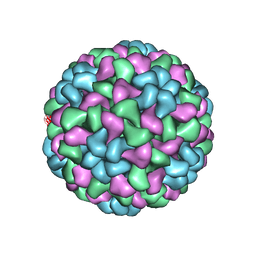 | |
1D6B
 
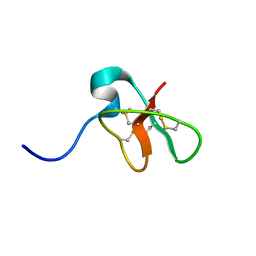 | | SOLUTION STRUCTURE OF DEFENSIN-LIKE PEPTIDE-2 (DLP-2) FROM PLATYPUS VENOM | | Descriptor: | DEFENSIN-LIKE PEPTIDE-2 | | Authors: | Torres, A.M, De Plater, G.M, Doverskog, M, C Birinyi-Strachan, L, Nicholson, G.M, Gallagher, C.H, Kuchel, P.W. | | Deposit date: | 1999-10-12 | | Release date: | 2000-06-21 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Defensin-like peptide-2 from platypus venom: member of a class of peptides with a distinct structural fold.
Biochem.J., 348, 2000
|
|
1DE3
 
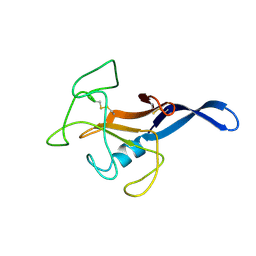 | | SOLUTION STRUCTURE OF THE CYTOTOXIC RIBONUCLEASE ALPHA-SARCIN | | Descriptor: | RIBONUCLEASE ALPHA-SARCIN | | Authors: | Perez-Canadillas, J.M, Campos-Olivas, R, Santoro, J, Lacadena, J, Martinez del Pozo, A, Gavilanes, J.G, Rico, M, Bruix, M. | | Deposit date: | 1999-11-12 | | Release date: | 2000-06-21 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The highly refined solution structure of the cytotoxic ribonuclease alpha-sarcin reveals the structural requirements for substrate recognition and ribonucleolytic activity.
J.Mol.Biol., 299, 2000
|
|
1QR5
 
 | | SOLUTION STRUCTURE OF HISTIDINE CONTAINING PROTEIN (HPR) FROM STAPHYLOCOCCUS CARNOSUS | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR | | Authors: | Kalbitzer, H.R, Gorler, A, Li, H, Dubovskii, P.V, Hengstenberg, W, Kowolik, C, Yamada, H, Akasaka, K. | | Deposit date: | 1999-05-19 | | Release date: | 2000-06-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | 15N and 1H NMR study of histidine containing protein (HPr) from Staphylococcus carnosus at high pressure.
Protein Sci., 9, 2000
|
|
1DM1
 
 | | 2.0 A CRYSTAL STRUCTURE OF THE DOUBLE MUTANT H(E7)V, T(E10)R OF MYOGLOBIN FROM APLYSIA LIMACINA | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Federici, L, Savino, C, Musto, R, Travaglini-Allocatelli, C, Cutruzzola, F, Brunori, M. | | Deposit date: | 1999-12-13 | | Release date: | 2000-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering His(E7) affects the control of heme reactivity in Aplysia limacina myoglobin.
Biochem.Biophys.Res.Commun., 269, 2000
|
|
