2KNT
 
 | |
2KSQ
 
 | | The myristoylated yeast ARF1 in a GTP and bicelle bound conformation | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-5'-TRIPHOSPHATE, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Liu, Y, Kahn, R, Prestegard, J. | | Deposit date: | 2010-01-12 | | Release date: | 2010-07-07 | | Last modified: | 2011-07-27 | | Method: | SOLUTION NMR | | Cite: | Dynamic structure of membrane-anchored Arf*GTP.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2KUC
 
 | | Solution Structure of a putative disulphide-isomerase from Bacteroides thetaiotaomicron | | Descriptor: | Putative disulphide-isomerase | | Authors: | Harris, R, Foti, R, Seidel, R.D, Bonanno, J.B, Freeman, J, Bain, K.T, Sauder, J.M, Burley, S.K, Girvin, M.E, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-17 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a putative disulphide-isomerase from Bacteroides thetaiotaomicron
To be Published
|
|
2KW4
 
 | | Solution NMR Structure of the Holo Form of a Ribonuclease H domain from D.hafniense, Northeast Structural Genomics Consortium Target DhR1A | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Mills, J.L, Eletsky, A, Hua, J, Belote, R.L, Buchwald, W.A, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target DhR1A
To be Published
|
|
2KYX
 
 | | Solution structure of the RRM domain of CYP33 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Park, S, Bushweller, J.H. | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The PHD3 domain of MLL acts as a CYP33-regulated switch between MLL-mediated activation and repression .
Biochemistry, 49, 2010
|
|
2KMB
 
 | |
2KMQ
 
 | |
2KNY
 
 | |
2KQP
 
 | |
2KRO
 
 | |
2KUM
 
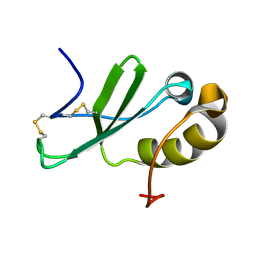 | | Solution structure of the human chemokine CCL27 | | Descriptor: | C-C motif chemokine 27 | | Authors: | Kirkpatrick, J.P, Jansma, A, Hsu, A, Handel, T.M, Nietlispach, D. | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR analysis of the structure, dynamics, and unique oligomerization properties of the chemokine CCL27.
J.Biol.Chem., 285, 2010
|
|
2KT4
 
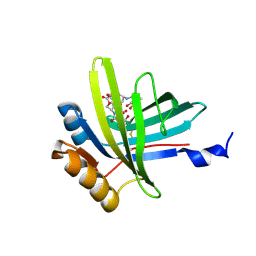 | | Lipocalin Q83 is a Siderocalin | | Descriptor: | Extracellular fatty acid-binding protein, GALLIUM (III) ION, N,N',N''-[(3S,7S,11S)-2,6,10-trioxo-1,5,9-trioxacyclododecane-3,7,11-triyl]tris(2,3-dihydroxybenzamide) | | Authors: | Coudevylle, N, Geist, L, Hartl, M, Kontaxis, G, Bister, K, Konrat, R. | | Deposit date: | 2010-01-18 | | Release date: | 2010-09-08 | | Last modified: | 2016-01-27 | | Method: | SOLUTION NMR | | Cite: | The v-myc-induced Q83 lipocalin is a siderocalin.
J.Biol.Chem., 285, 2010
|
|
2KVI
 
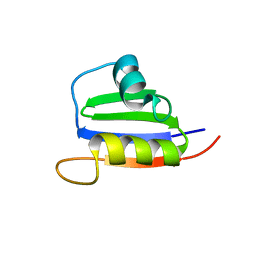 | | Structure of Nab3 RRM | | Descriptor: | Nuclear polyadenylated RNA-binding protein 3 | | Authors: | Pergoli, R, Kubicek, K, Hobor, F, Pasulka, J, Stefl, R. | | Deposit date: | 2010-03-15 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and RNA-binding study of RNA-recognition motif of Nab3
J.Biol.Chem., 2010
|
|
2KTY
 
 | |
2KVF
 
 | |
2KQ2
 
 | | Solution NMR structure of the apo form of a ribonuclease H domain of protein DSY1790 from Desulfitobacterium hafniense, Northeast Structural Genomics target DhR1A | | Descriptor: | Ribonuclease H-related protein | | Authors: | Mills, J.L, Eletsky, A, Hua, J, Belote, R.L, Buchwald, W.A, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-24 | | Release date: | 2010-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the apo form of a ribonuclease H domain of protein DSY1790 from Desulfitobacterium hafniense, Northeast Structural Genomics target DhR1A
To be Published
|
|
2KTD
 
 | | Solution structure of mouse lipocalin-type prostaglandin D synthase / substrate analog (U-46619) complex | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, Prostaglandin-H2 D-isomerase | | Authors: | Shimamoto, S, Maruo, H, Yoshida, T, Kato, N, Ohkubo, T. | | Deposit date: | 2010-01-27 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Lipocalin-type Prostaglandin D synthase / Substrate analog complex reveals Open-Closed Conformational Change required for Substrate Recognition
To be Published
|
|
2OHP
 
 | | X-ray crystal structure of beta secretase complexed with compound 3 | | Descriptor: | 6-[2-(1H-INDOL-6-YL)ETHYL]PYRIDIN-2-AMINE, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Patel, S. | | Deposit date: | 2007-01-10 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Application of fragment screening by X-ray crystallography to the discovery of aminopyridines as inhibitors of beta-secretase.
J.Med.Chem., 50, 2007
|
|
2NUZ
 
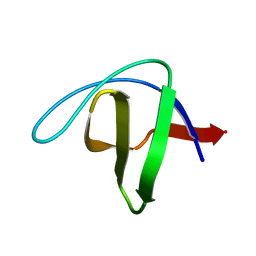 | | crystal structure of alpha spectrin SH3 domain measured at room temperature | | Descriptor: | Spectrin alpha chain, brain | | Authors: | Agarwal, V, Faelber, K, Hologne, M, Chevelkov, V, Oschkinat, H, Diehl, A, Reif, B. | | Deposit date: | 2006-11-10 | | Release date: | 2007-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparison of MAS solid-state NMR and X-ray crystallographic data in the analysis of protein dynamics in the solid state
To be Published
|
|
2NZ5
 
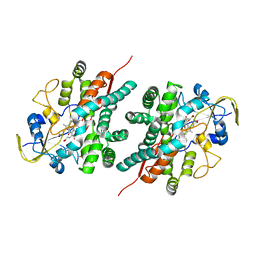 | | Structure and Function Studies of Cytochrome P450 158A1 from Streptomyces coelicolor A3(2) | | Descriptor: | Cytochrome P450 CYP158A1, PROTOPORPHYRIN IX CONTAINING FE, naphthalene-1,2,4,5,7-pentol | | Authors: | Zhao, B, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2006-11-22 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Different binding modes of two flaviolin substrate molecules in cytochrome P450 158A1 (CYP158A1) compared to CYP158A2.
Biochemistry, 46, 2007
|
|
2NT3
 
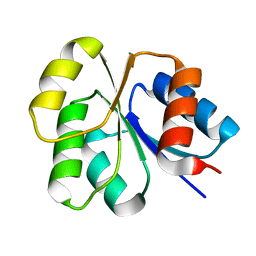 | | Receiver domain from Myxococcus xanthus social motility protein FrzS (Y102A Mutant) | | Descriptor: | Response regulator homolog | | Authors: | Fraser, J.S, Echols, N, Merlie, J.P, Zusman, D.R, Alber, T. | | Deposit date: | 2006-11-06 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS.
Mol.Microbiol., 65, 2007
|
|
2O2W
 
 | |
2O31
 
 | |
2NX0
 
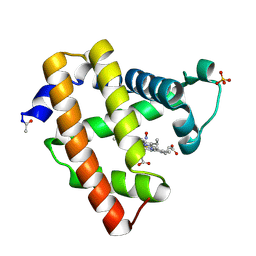 | | Ferrous nitrosyl blackfin tuna myoglobin | | Descriptor: | Myoglobin, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Schreiter, E.R, Rodriguez, M.M, Weichsel, A, Montfort, W.R, Bonaventura, J. | | Deposit date: | 2006-11-16 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | S-nitrosylation-induced conformational change in blackfin tuna myoglobin.
J.Biol.Chem., 282, 2007
|
|
2O85
 
 | | S. Aureus thioredoxin P31T mutant | | Descriptor: | Thioredoxin | | Authors: | Roos, G, Loris, R, Messens, J. | | Deposit date: | 2006-12-12 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The conserved active site proline determines the reducing power of Staphylococcus aureus thioredoxin
J.Mol.Biol., 368, 2007
|
|
