6UEF
 
 | | Crystal Structure of a Self-Assembling DNA Scaffold Containing Sequence Modifications to Attempt to Disrupt Crystal Contacts in a Rhombohedral Lattice. | | Descriptor: | DNA (5'-D(*CP*AP*AP*TP*GP*AP*CP*TP*CP*AP*TP*GP*CP*TP*CP*AP*TP*CP*GP*GP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*GP*AP*TP*G)-3'), DNA (5'-D(P*AP*GP*CP*AP*TP*GP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A Self-Assembled Rhombohedral DNA Crystal Scaffold with Tunable Cavity Sizes and High-Resolution Structural Detail.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
9EOQ
 
 | | Cryo-EM Structure of a 1033 Scaffold Base DNA Origami Nanostructure V4 and TBA | | Descriptor: | DNA (42-MER), DNA (5'-D(P*AP*TP*AP*TP*AP*GP*CP*GP*TP*GP*GP*AP*AP*GP*T)-3') | | Authors: | Ali, K, Georg, K, Volodymyr, M, Johanna, G, Maximilian, N.H, Lukas, K, Simone, C, Hendrik, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Designing Rigid DNA Origami Templates for Molecular Visualization Using Cryo-EM.
Nano Lett., 24, 2024
|
|
7B72
 
 | | DNA duplex with phosphoryl guanidine moiety, Rp-diastereomer | | Descriptor: | DNA (5'-D(*CP*AP*CP*GP*CP*CP*GP*CP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*CP*GP*GP*CP*GP*(RGT)P*G)-3') | | Authors: | Lomzov, A.A, Pyshnyi, D.V, Shernuykov, A.V, Apukhtina, V.S. | | Deposit date: | 2020-12-09 | | Release date: | 2021-01-13 | | Last modified: | 2023-08-23 | | Method: | SOLUTION NMR | | Cite: | DNA duplex with phosphoryl guanidine moiety, Rp-diastereomer
To Be Published
|
|
6TZS
 
 | | A DNA i-motif/duplex hybrid | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*CP*TP*GP*(CBR)P*AP*A)-3') | | Authors: | Chu, B, Paukstelis, P.J. | | Deposit date: | 2019-08-13 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A DNA G-quadruplex/i-motif hybrid.
Nucleic Acids Res., 47, 2019
|
|
4YS5
 
 | |
5TGP
 
 | | DNA 8mer containing two 2SeT modifications | | Descriptor: | DNA/RNA (5'-R(*G)-D(P*(2ST))-R(P*G)-D(P*(2ST))-R(P*AP*CP*AP*C)-3') | | Authors: | Zhang, W, Huang, Z. | | Deposit date: | 2016-09-28 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA 8mer containing two 2SeT modifications
To Be Published
|
|
3UBI
 
 | | The Absence of Tertiary Interactions in a Self-Assembled DNA Crystal Structure | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*CP*GP*TP*AP*CP*TP*CP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*AP*GP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*CP*GP*AP*GP*TP*AP*CP*GP*AP*CP*GP*AP*CP*AP*AP*G)-3'), ... | | Authors: | Nguyen, N, Birktoft, J.J, Sha, R, Wang, T, Zheng, J, Constantinou, P.E, Ginell, S.L, Chen, Y, Mao, C, Seeman, N.C. | | Deposit date: | 2011-10-24 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (6.8046 Å) | | Cite: | The absence of tertiary interactions in a self-assembled DNA crystal structure.
J.Mol.Recognit., 25, 2012
|
|
8PJL
 
 | |
5BZY
 
 | |
5BZ7
 
 | |
5BXW
 
 | |
5BZ9
 
 | |
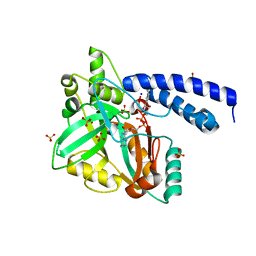
1TAE
 
 | |
287D
 
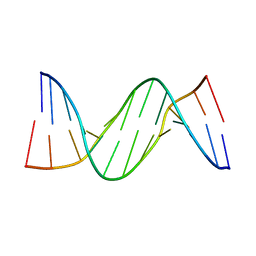 | |
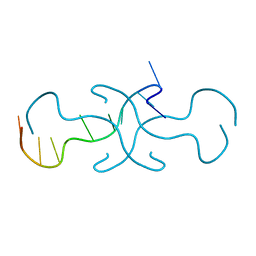
4RIP
 
 | |
297D
 
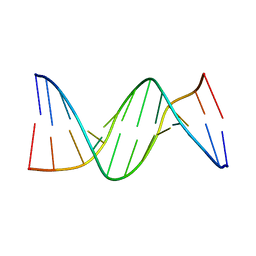 | |
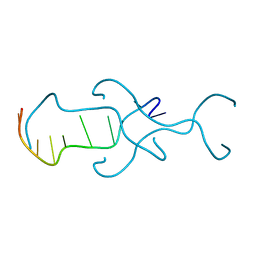
4RIM
 
 | |
1JIH
 
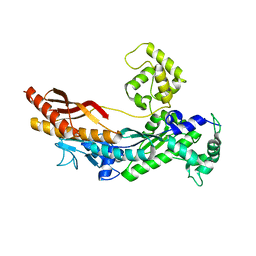 | | Yeast DNA Polymerase ETA | | Descriptor: | DNA Polymerase ETA | | Authors: | Trincao, J, Johnson, R.E, Escalante, C.R, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2001-07-02 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis
Mol.Cell, 8, 2001
|
|
4HTP
 
 | |
4HTO
 
 | |
2M44
 
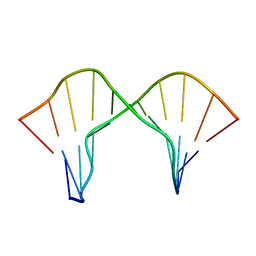 | | DNA containing a cluster of 8-oxo-guanine and abasic site lesion: beta anomer (6AP, 8OG14) | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*CP*(AAB)P*CP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*(8OG)P*TP*GP*GP*GP*AP*GP*CP*G)-3') | | Authors: | Zalesak, J, Jourdan, M, Constant, J, Lourdin, M. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of DNA duplexes containing a cluster of mutagenic 8-oxoguanine and abasic site lesions.
J.Mol.Biol., 426, 2014
|
|
2M3Y
 
 | | DNA containing a cluster of 8-oxo-guanine and abasic site lesion: beta anomer | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*CP*(AAB)P*CP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*GP*GP*AP*(8OG)P*CP*G)-3') | | Authors: | Zalesak, J, Jourdan, M, Constant, J, Lourdin, M. | | Deposit date: | 2013-01-28 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of DNA duplexes containing a cluster of mutagenic 8-oxoguanine and abasic site lesions.
J.Mol.Biol., 426, 2014
|
|
2M43
 
 | | DNA containing a cluster of 8-oxo-guanine and abasic site lesion: alpha anomer (AP6, 8OG 14) | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*CP*(ORP)P*CP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*(8OG)P*TP*GP*GP*GP*AP*GP*CP*G)-3') | | Authors: | Zalesak, J, Jourdan, M, Constant, J, Lourdin, M. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of DNA duplexes containing a cluster of mutagenic 8-oxoguanine and abasic site lesions.
J.Mol.Biol., 426, 2014
|
|
2M3P
 
 | | DNA containing a cluster of 8-oxo-guanine and abasic site lesion: alpha anomer | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*CP*(ORP)P*CP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*GP*GP*AP*(8OG)P*CP*G)-3') | | Authors: | Zalesak, J, Jourdan, M, Lourdin, M, Constant, J. | | Deposit date: | 2013-01-25 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of DNA duplexes containing a cluster of mutagenic 8-oxoguanine and abasic site lesions.
J.Mol.Biol., 426, 2014
|
|
2M40
 
 | | DNA containing a cluster of 8-oxo-guanine and THF lesion | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*CP*(3DR)P*CP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*GP*GP*AP*(8OG)P*CP*G)-3') | | Authors: | Zalesak, J, Jourdan, M, Constant, J, Lourdin, M. | | Deposit date: | 2013-01-28 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of DNA duplexes containing a cluster of mutagenic 8-oxoguanine and abasic site lesions.
J.Mol.Biol., 426, 2014
|
|
