6MN0
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, H168A mutant in complex with acetyl-CoA | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETYL COENZYME *A, Aminoglycoside N(3)-acetyltransferase, ... | | Authors: | Stogios, P.J, Skarina, T, Zu, X, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
3SA2
 
 | |
6Y93
 
 | | Crystal structure of the DNA-binding domain of the Nucleoid Occlusion Factor (Noc) complexed to the Noc-binding site (NBS) | | Descriptor: | Noc Binding Site (NBS), Nucleoid occlusion protein | | Authors: | Jalal, A.S.B, Tran, N.T, Stevenson, C.E.M, Chan, E, Lo, R, Tan, X, Noy, A, Lawson, D.M, Le, T.B.K. | | Deposit date: | 2020-03-06 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Diversification of DNA-Binding Specificity by Permissive and Specificity-Switching Mutations in the ParB/Noc Protein Family.
Cell Rep, 32, 2020
|
|
3SGI
 
 | |
6NAE
 
 | |
6ZL5
 
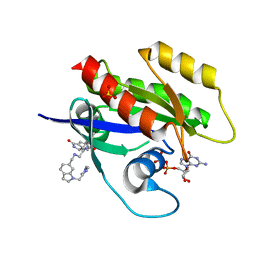 | | CRYSTAL STRUCTURE OF KRAS-G12D(C118S) IN COMPLEX WITH BI-2852 AND GDP | | Descriptor: | (3~{S})-3-[2-[[[1-[(1-methylimidazol-4-yl)methyl]indol-6-yl]methylamino]methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kessler, D, Fischer, G, Boettcher, J. | | Deposit date: | 2020-06-30 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.645 Å) | | Cite: | Drugging all RAS isoforms with one pocket.
Future Med Chem, 12, 2020
|
|
6ZIR
 
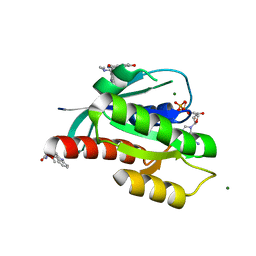 | | CRYSTAL STRUCTURE OF NRAS (C118S) IN COMPLEX WITH GDP AND COMPOUND 18 | | Descriptor: | (3~{S})-3-[2-[(dimethylamino)methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase NRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kessler, D, Fischer, G, Boettcher, J. | | Deposit date: | 2020-06-26 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Drugging all RAS isoforms with one pocket.
Future Med Chem, 12, 2020
|
|
3T0J
 
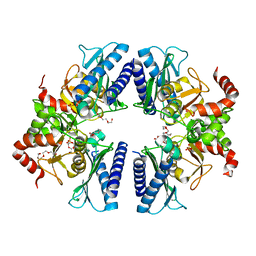 | |
6ZL3
 
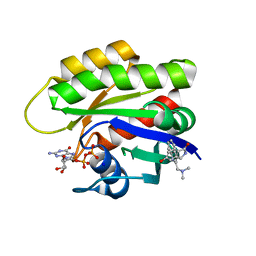 | | CRYSTAL STRUCTURE OF HRAS IN COMPLEX WITH COMPOUND 18 and GDP | | Descriptor: | (3~{S})-3-[2-[(dimethylamino)methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kessler, D, Fischer, G, Boettcher, J. | | Deposit date: | 2020-06-30 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Drugging all RAS isoforms with one pocket.
Future Med Chem, 12, 2020
|
|
3T9C
 
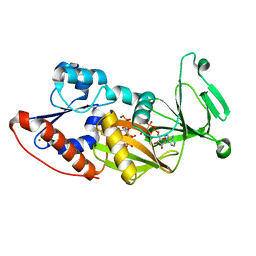 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMPPNP and inositol hexakisphosphate (IP6) | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Inositol Pyrophosphate Kinase, MAGNESIUM ION, ... | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
6NTI
 
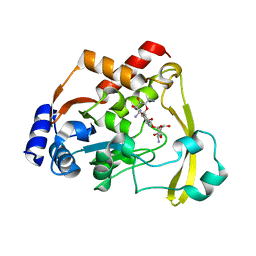 | | Neutron/X-ray crystal structure of AAC-VIa bound to kanamycin b | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, Aminoglycoside N(3)-acetyltransferase, MAGNESIUM ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2019-01-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2.3 Å), X-RAY DIFFRACTION | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6C7N
 
 | | Monoclinic form of malic enzyme from sorghum at 2 angstroms resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Malic enzyme, ... | | Authors: | Trajtenberg, F, Alvarez, C, Buschiazzo, A. | | Deposit date: | 2018-01-23 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular adaptations of NADP-malic enzyme for its function in C4photosynthesis in grasses.
Nat.Plants, 5, 2019
|
|
6Z1X
 
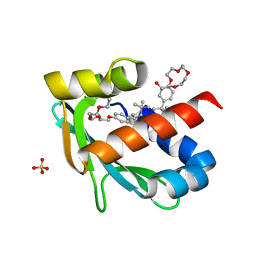 | |
6Z1W
 
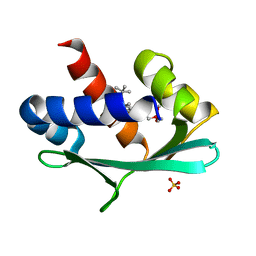 | |
6YWI
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148E variant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
6YWH
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148D variant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, ... | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
6YWQ
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148H variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
6YXC
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148R variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, SODIUM ION | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
6YWR
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148H variant (space group C2) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, SULFATE ION | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
6YWG
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148N variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into the mechanisms of light-oxygen-voltage domain color tuning from a set of high-resolution X-ray structures.
Proteins, 2021
|
|
2GUU
 
 | | crystal structure of Plasmodium vivax orotidine 5-monophosphate decarboxylase with 6-aza-UMP bound | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, ODcase | | Authors: | Dong, A, Lew, J, Zhao, Y, Sundararajan, E, Wasney, G, Vedadi, M, Koeieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Pai, E.F, Kotra, L, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-01 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of Plasmodium vivax orotidine 5-monophosphate decarboxylase with 6-aza-UMP bound
To be Published
|
|
6YWA
 
 | |
6YWB
 
 | |
6MCJ
 
 | |
4Z27
 
 | |
