5OJZ
 
 | |
5O6P
 
 | |
7RVB
 
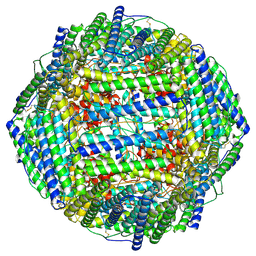 | | High resolution map of molecular chaperone Artemin | | Descriptor: | Ferritin | | Authors: | Parvate, A.D, Powell, S.M, Brookreason, J.T, Novikova, I.V, Evans, J.E. | | Deposit date: | 2021-08-18 | | Release date: | 2022-12-14 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.04 Å) | | Cite: | Cryo-EM structure of the diapause chaperone artemin.
Front Mol Biosci, 9, 2022
|
|
1I6B
 
 | |
8FA1
 
 | |
5HJF
 
 | |
7TIK
 
 | |
3IXR
 
 | | Crystal Structure of Xylella fastidiosa PrxQ C47S Mutant | | Descriptor: | Bacterioferritin comigratory protein | | Authors: | Horta, B.B, Oliveira, M.A, Discola, K.F, Cussiol, J.R.R, Netto, L.E.S. | | Deposit date: | 2009-09-04 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and Structural Analysis Indicates that PrxQ, a Cys Based Peroxidase from Xylella fastidiosa, possesses high reactivity towards hydroperoxides
To be Published
|
|
5IPG
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure FFT-butyl (Hyperoxodized by t-butyl hydroperoxide) | | Descriptor: | Bacterioferritin comigratory protein, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-09 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IOX
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure LUss | | Descriptor: | Bacterioferritin comigratory protein | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-09 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IMV
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F6 | | Descriptor: | Bacterioferritin comigratory protein, FORMIC ACID, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-07 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IMA
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F2 | | Descriptor: | Bacterioferritin comigratory protein, FORMIC ACID, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-06 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IMF
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F5 | | Descriptor: | Bacterioferritin comigratory protein, FORMIC ACID, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-06 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IPH
 
 | | Xanthomonas campestris Peroxiredoxin Q - C84S mutant | | Descriptor: | Bacterioferritin comigratory protein, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-09 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IMD
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F4 | | Descriptor: | Bacterioferritin comigratory protein, FORMIC ACID, OXYGEN ATOM, ... | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-06 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5INY
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F8 | | Descriptor: | Bacterioferritin comigratory protein, FORMIC ACID, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IIZ
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F0 | | Descriptor: | Bacterioferritin comigratory protein, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-01 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IMC
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F3 | | Descriptor: | Bacterioferritin comigratory protein, FORMIC ACID, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-06 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IO0
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F9 | | Descriptor: | Bacterioferritin comigratory protein, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IOW
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure FFcumene (Hyperoxidized by cumene hydroperoxide) | | Descriptor: | Bacterioferritin comigratory protein, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-09 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IM9
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F1 | | Descriptor: | Bacterioferritin comigratory protein, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-05 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IMZ
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F7 | | Descriptor: | Bacterioferritin comigratory protein, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-07 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IO2
 
 | | Xanthomonas campestris Peroxiredoxin Q - C48S mutant | | Descriptor: | Bacterioferritin comigratory protein, PHOSPHATE ION, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
2J4U
 
 | |
2MD4
 
 | |
