2MVK
 
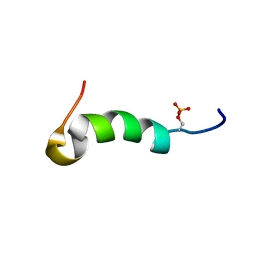 | | Solution structure of phosphorylated cytosolic part of Trop2 | | Descriptor: | Tumor-associated calcium signal transducer 2 | | Authors: | Ilc, G, Plavec, J, Vidmar, T, Pavsic, M, Lenarcic, B. | | Deposit date: | 2014-10-08 | | Release date: | 2015-05-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The cytosolic tail of the tumor marker protein Trop2--a structural switch triggered by phosphorylation.
Sci Rep, 5, 2015
|
|
2MWR
 
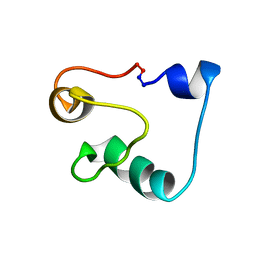 | | Solution Structure of Acidocin B, a Circular Bacteriocin from Lactobacillus acidophilus M46 | | Descriptor: | Acidocin B | | Authors: | Vederas, J.C, Acedo, J.Z, van Belkum, M.J, Lohans, C.T. | | Deposit date: | 2014-11-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Acidocin B, a Circular Bacteriocin Produced by Lactobacillus acidophilus M46.
Appl.Environ.Microbiol., 81, 2015
|
|
2N0M
 
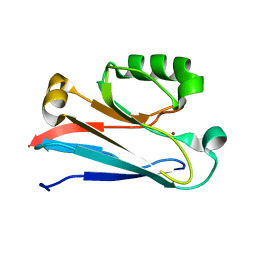 | | The solution structure of the soluble form of the Lipid-modified Azurin from Neisseria gonorrhoeae | | Descriptor: | COPPER (I) ION, Lipid modified azurin protein | | Authors: | Pauleta, S.R, Matzapetakis, M.F, Nobrega, C.F, Carreira, C, Saraiva, I.H. | | Deposit date: | 2015-03-10 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the soluble form of the lipid-modified azurin from Neisseria gonorrhoeae, the electron donor of cytochrome c peroxidase.
Biochim.Biophys.Acta, 1857, 2016
|
|
2MVL
 
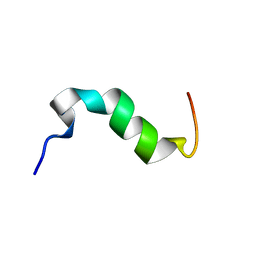 | | Solution structure of cytosolic part of Trop2 | | Descriptor: | Tumor-associated calcium signal transducer 2 | | Authors: | Ilc, G, Plavec, J, Vidmar, T, Pavsic, M, Lenarcic, B. | | Deposit date: | 2014-10-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The cytosolic tail of the tumor marker protein Trop2--a structural switch triggered by phosphorylation.
Sci Rep, 5, 2015
|
|
2KEL
 
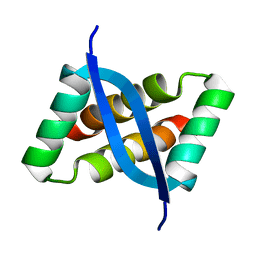 | | Structure of the transcription regulator SvtR from the hyperthermophilic archaeal virus SIRV1 | | Descriptor: | Uncharacterized protein 56B | | Authors: | Guilliere, F, Kessler, A, Peixeiro, N, Sezonov, G, Prangishvili, D, Delepierre, M, Guijarro, J.I. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, function, and targets of the transcriptional regulator SvtR from the hyperthermophilic archaeal virus SIRV1.
J.Biol.Chem., 284, 2009
|
|
2ARI
 
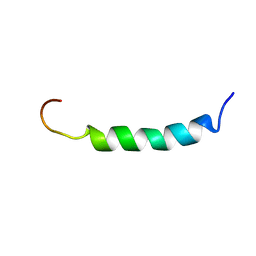 | | Solution structure of micelle-bound fusion domain of HIV-1 gp41 | | Descriptor: | Envelope polyprotein GP160 | | Authors: | Jaroniec, C.P, Kaufman, J.D, Stahl, S.J, Viard, M, Blumenthal, R, Wingfield, P.T, Bax, A. | | Deposit date: | 2005-08-19 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of Micelle-Associated Human Immunodeficiency Virus gp41 Fusion Domain.
Biochemistry, 44, 2005
|
|
2NS4
 
 | |
2HTF
 
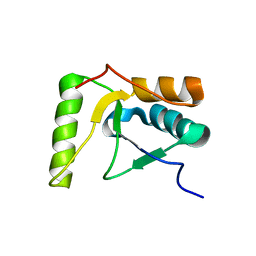 | | The solution structure of the BRCT domain from human polymerase reveals homology with the TdT BRCT domain | | Descriptor: | DNA polymerase mu | | Authors: | DeRose, E.F, Clarkson, M.W, Gilmore, S.A, Ramsden, D.A, Mueller, G.A, London, R.E, Lee, A.L. | | Deposit date: | 2006-07-25 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of polymerase mu's BRCT Domain reveals an element essential for its role in nonhomologous end joining.
Biochemistry, 46, 2007
|
|
2NAR
 
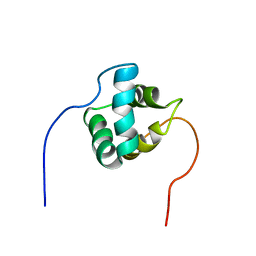 | | Solution structure of AVR3a_60-147 from Phytophthora infestans | | Descriptor: | Effector protein Avr3a | | Authors: | Matena, A, Bayer, P, Zhukov, I, Stanek, J, Kozminski, W, van West, P, Wawra, S. | | Deposit date: | 2016-01-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The RxLR Motif of the Host Targeting Effector AVR3a ofPhytophthora infestansIs Cleaved before Secretion.
Plant Cell, 29, 2017
|
|
2N8Z
 
 | | Apo form of Calmodulin-Like Domain of Human Non-Muscle alpha-actinin 1 | | Descriptor: | Alpha-actinin-1 | | Authors: | Drmota Prebil, S, Slapsak, U, de Almeida Ribeiro, E, Pavsic, M, Ilc, G, Zielinska, K, Hartl, M, Backman, L, Plavec, J, Lenarcic, B, Djinovic-Carugo, K. | | Deposit date: | 2015-10-28 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and calcium-binding studies of calmodulin-like domain of human non-muscle alpha-actinin-1.
Sci Rep, 6, 2016
|
|
2LS0
 
 | |
2N8Y
 
 | | Holo form of Calmodulin-Like Domain of Human Non-Muscle alpha-actinin 1 | | Descriptor: | Alpha-actinin-1, CALCIUM ION | | Authors: | Drmota Prebil, S, Slapsak, U, de Almeida Ribeiro, E, Pavsic, M, Ilc, G, Zielinska, K, Hartl, M, Backman, L, Plavec, J, Lenarcic, B, Djinovic-Carugo, K. | | Deposit date: | 2015-10-28 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and calcium-binding studies of calmodulin-like domain of human non-muscle alpha-actinin-1.
Sci Rep, 6, 2016
|
|
2JVY
 
 | | Solution Structure of the EDA-ID-related C417F mutant of human NEMO zinc finger | | Descriptor: | NF-kappa-B essential modulator, ZINC ION | | Authors: | Cordier, F, Vinolo, E, Veron, M, Delepierre, M, Agou, F. | | Deposit date: | 2007-09-28 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of NEMO zinc finger and impact of an anhidrotic ectodermal dysplasia with immunodeficiency-related point mutation.
J.Mol.Biol., 377, 2008
|
|
2MVS
 
 | | N6-Methyladenosine RNA | | Descriptor: | N-6_Methyl_Adenosine_RNA | | Authors: | Lynch, S.R, Kool, E. | | Deposit date: | 2014-10-10 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification.
J.Am.Chem.Soc., 137, 2015
|
|
2MVY
 
 | |
2N5F
 
 | |
2LTJ
 
 | | Conformational analysis of StrH, the surface-attached exo- beta-D-N-acetylglucosaminidase from Streptococcus pneumoniae | | Descriptor: | Beta-N-acetylhexosaminidase | | Authors: | Pluvinage, B, Chitayat, S, Ficko-Blean, E, Abbott, D, Kunjachen, J, Grondin, J, Spencer, H, Smith, S, Boraston, A. | | Deposit date: | 2012-05-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational analysis of StrH, the surface-attached exo-beta-D-N-acetylglucosaminidase from Streptococcus pneumoniae.
J.Mol.Biol., 425, 2013
|
|
2LPB
 
 | | Structure of the complex of the central activation domain of Gcn4 bound to the mediator co-activator domain 1 of Gal11/med15 | | Descriptor: | General control protein GCN4, Mediator of RNA polymerase II transcription subunit 15 | | Authors: | Brzovic, P.S, Heikaus, C.C, Kisselev, L, Vernon, R, Herbig, E, Pacheco, D, Warfield, L, Littlefield, P, Baker, D, Klevit, R.E, Hahn, S. | | Deposit date: | 2012-02-07 | | Release date: | 2012-02-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex.
Mol.Cell, 44, 2011
|
|
2LGY
 
 | | Ubiquitin-like domain from HOIL-1 | | Descriptor: | RanBP-type and C3HC4-type zinc finger-containing protein 1 | | Authors: | Beasley, S.A, Shaw, G.S. | | Deposit date: | 2011-08-03 | | Release date: | 2012-06-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the E3 ligase HOIL-1 Ubl domain.
Protein Sci., 21, 2012
|
|
2JMW
 
 | | Structure of DNA-Binding Domain of Arabidopsis GT-1 | | Descriptor: | DNA binding protein GT-1 | | Authors: | Nagata, T, Niyada, E, Noto, K, Ikeda, Y, Yamamoto, Y, Uesugi, S, Murata, J, Hiratsuka, K, Katahira, M. | | Deposit date: | 2006-12-11 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: mechanism for an increase in DNA-binding affinity through phosphorylation.
Proteins, 78, 2010
|
|
2KI7
 
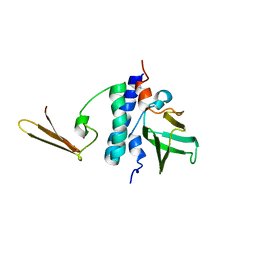 | | The solution structure of RPP29-RPP21 complex from Pyrococcus furiosus | | Descriptor: | Ribonuclease P protein component 1, Ribonuclease P protein component 4, ZINC ION | | Authors: | Xu, Y, Foster, M.P. | | Deposit date: | 2009-04-28 | | Release date: | 2009-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an archaeal RNase P binary protein complex: formation of the 30-kDa complex between Pyrococcus furiosus RPP21 and RPP29 is accompanied by coupled protein folding and highlights critical features for protein-protein and protein-RNA interactions.
J.Mol.Biol., 393, 2009
|
|
2IHX
 
 | |
2PAS
 
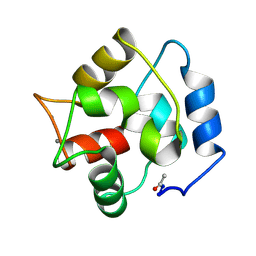 | | COMPARISON BETWEEN THE CRYSTAL AND THE SOLUTION STRUCTURES OF THE EF HAND PARVALBUMIN | | Descriptor: | CALCIUM ION, PARVALBUMIN | | Authors: | Padilla, A, Cave, A, Parello, J, Etienne, G, Baldellon, C. | | Deposit date: | 1994-03-22 | | Release date: | 1994-06-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Comparison between the Crystal and the Solution Structures of the EF Hand Parvalbumin
To be Published
|
|
2L68
 
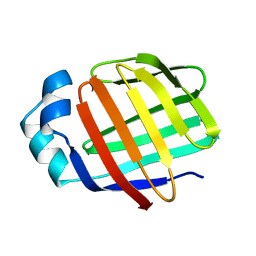 | | Solution Structure of Human Holo L-FABP | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Cai, J, Luecke, C, Chen, Z, Qiao, Y, Klimtchuk, E.S, Hamilton, J.A. | | Deposit date: | 2010-11-17 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited.
Biophys.J., 102, 2012
|
|
2L67
 
 | | Solution Structure of Human Apo L-FABP | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Cai, J, Luecke, C, Chen, Z, Qiao, Y, Klimtchuk, E.S, Hamilton, J.A. | | Deposit date: | 2010-11-17 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited.
Biophys.J., 102, 2012
|
|
