7YDL
 
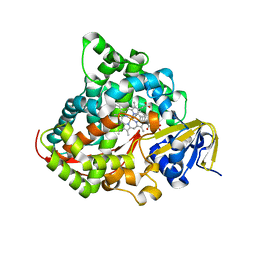 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268I/A184V/A82T in complex with N-imidazolyl-hexanoyl-L-phenylalanine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A/T268I/A184V/A82T in complex with N-imidazolyl-hexanoyl-L-phenylalanine
To Be Published
|
|
7YDE
 
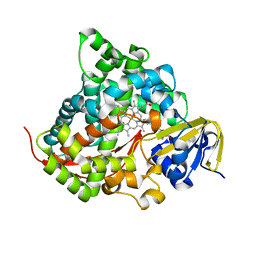 | | Crystal structure of the P450 BM3 heme domain mutant F87T/T268V/I263V in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-04 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YDD
 
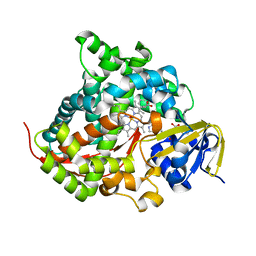 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268P/V78I in complex with N-imidazolyl-pentanoyl-L-phenylalanine,propylbenzene and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, HYDROXYAMINE, P450 BM3 heme domain mutant F87A/T268P/V78I, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-04 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.663 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YDC
 
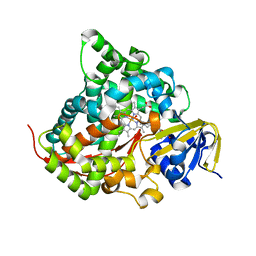 | | Crystal structure of the P450 BM3 heme domain mutant F87L/T268V/V78C in complex with N-imidazolyl-pentanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-04 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.609 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YDB
 
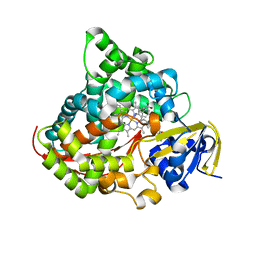 | | Crystal structure of the P450 BM3 heme domain mutant F87V/T268I in complex with N-imidazolyl-pentanoyl-L-phenylalanine,ethylbenzene and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-04 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.472 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YDA
 
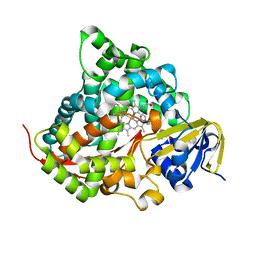 | | Crystal structure of the P450 BM3 heme domain mutant F87V/T268V/A184V in complex with N-imidazolyl-pentanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-04 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.557 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YD9
 
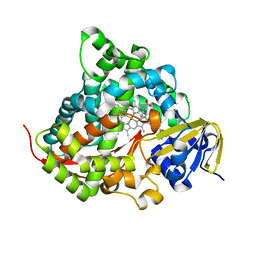 | | Crystal structure of the P450 BM3 heme domain mutant F87G/T268V/A184V/A328V in complex with N-imidazolyl-hexanoyl-L-phenylalanine,methylbenzene and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-04 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YD8
 
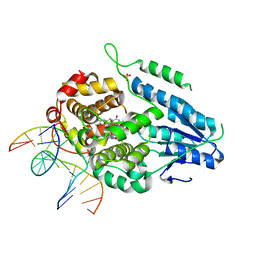 | | TR-SFX MmCPDII-DNA complex: 2 ns snapshot. Includes 2 ns, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YD7
 
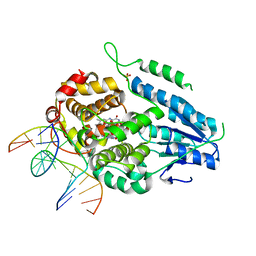 | | TR-SFX MmCPDII-DNA complex: 1 ns snapshot. Includes 1 ns, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YD6
 
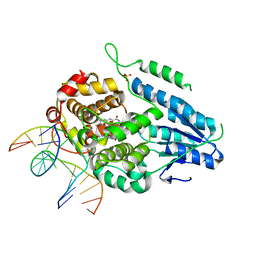 | | TR-SFX MmCPDII-DNA complex: 650 ps snapshot. Includes 650ps, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YD4
 
 | | Crystal structure of an N terminal truncated secreted protein, Rv0398c from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, Possible secreted protein | | Authors: | Saha, R, Mukherjee, S, Singh, B.K, Weiss, M.S, De, S, Das, A.K. | | Deposit date: | 2022-07-03 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Crystal structure of a mycobacterial secretory protein Rv0398c and in silico prediction of its export pathway.
Biochem.Biophys.Res.Commun., 672, 2023
|
|
7YD3
 
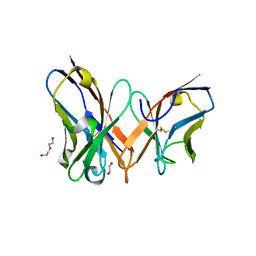 | | Single-chain variable fragment of app 3D1 antibody | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yan, L, Yang, G. | | Deposit date: | 2022-07-03 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Cross-reactive epitopes between HIV and Coronavirus revealed by 3D1
To Be Published
|
|
7YD2
 
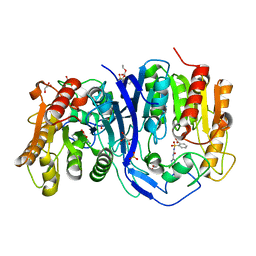 | | SulE_P44R_S209A | | Descriptor: | 2-[(4-chloranyl-6-methoxy-pyrimidin-2-yl)carbamoylsulfamoyl]benzoic acid, 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, Alpha/beta fold hydrolase, ... | | Authors: | Liu, B, Ran, T, Wang, W, He, J. | | Deposit date: | 2022-07-03 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
7YCT
 
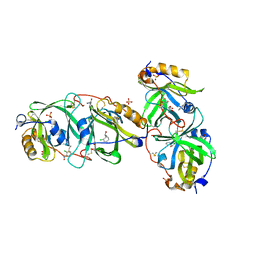 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE, Oxidus gracilis complexed with (R)-2-Chloromandelonitrile | | Descriptor: | (2~{R})-2-(2-chlorophenyl)-2-oxidanyl-ethanenitrile, GLYCEROL, Hydroxynitrile lyase, ... | | Authors: | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | Deposit date: | 2022-07-01 | | Release date: | 2024-01-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Based Site-Directed Mutagenesis of Hydroxynitrile Lyase from Cyanogenic Millipede, Oxidus gracilis for Hydrocyanation and Henry Reactions.
Chembiochem, 25, 2024
|
|
7YCR
 
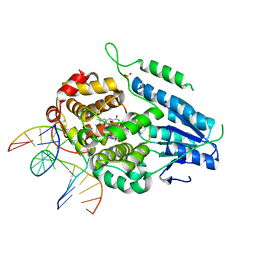 | | TR-SFX MmCPDII-DNA complex: 450 ps snapshot. Includes 450ps, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YCP
 
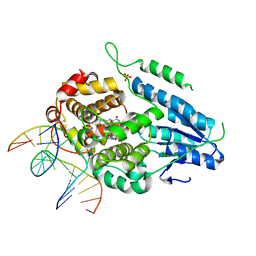 | | TR-SFX MmCPDII-DNA complex: 250 ps snapshot. Includes 250 ps, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YCN
 
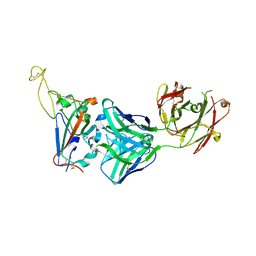 | |
7YCM
 
 | | TR-SFX MmCPDII-DNA complex: 100 ps snapshot. Includes 100ps, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YCL
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with IS-9A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IS-9A Fab heavy chain, IS-9A Fab light chain, ... | | Authors: | Mohapatra, A, Chen, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7YCK
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with FP-12A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, FP-12A Fab heavy chain, FP-12A Fab light chain, ... | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7YCF
 
 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE, Oxidus gracilis IN ACETONITRILE | | Descriptor: | 2-HYDROXY-2-METHYLPROPANENITRILE, CHLORIDE ION, Hydroxynitrile lyase, ... | | Authors: | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | Deposit date: | 2022-07-01 | | Release date: | 2024-01-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Based Site-Directed Mutagenesis of Hydroxynitrile Lyase from Cyanogenic Millipede, Oxidus gracilis for Hydrocyanation and Henry Reactions.
Chembiochem, 25, 2024
|
|
7YCE
 
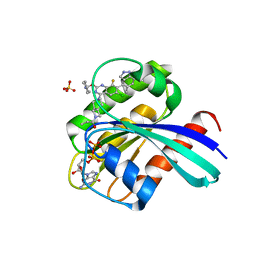 | | KRas G12C in complex with Compound 7b | | Descriptor: | 1-[7-[6-chloranyl-2-(1-ethylpiperidin-4-yl)oxy-8-fluoranyl-7-(5-methyl-1~{H}-indazol-4-yl)quinazolin-4-yl]-2,7-diazaspiro[3.5]nonan-2-yl]propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Amano, Y. | | Deposit date: | 2022-07-01 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and biological evaluation of 1-{2,7-diazaspiro[3.5]nonan-2-yl}prop-2-en-1-one derivatives as covalent inhibitors of KRAS G12C with favorable metabolic stability and anti-tumor activity.
Bioorg.Med.Chem., 71, 2022
|
|
7YCD
 
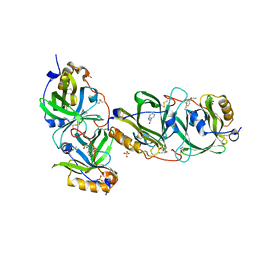 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE, Oxidus gracilis bound with (R)-(+)-ALPHA-HYDROXYBENZENE-ACETONITRILE | | Descriptor: | (2R)-hydroxy(phenyl)ethanenitrile, Hydroxynitrile lyase, SULFATE ION | | Authors: | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | Deposit date: | 2022-07-01 | | Release date: | 2024-01-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Based Site-Directed Mutagenesis of Hydroxynitrile Lyase from Cyanogenic Millipede, Oxidus gracilis for Hydrocyanation and Henry Reactions.
Chembiochem, 25, 2024
|
|
7YCC
 
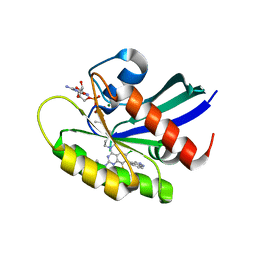 | | KRas G12C in complex with Compound 5c | | Descriptor: | 1-[7-[6-chloranyl-8-fluoranyl-7-(5-methyl-1~{H}-indazol-4-yl)-2-[(1-methylpiperidin-4-yl)amino]quinazolin-4-yl]-2,7-diazaspiro[3.5]nonan-2-yl]propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Amano, Y. | | Deposit date: | 2022-07-01 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery and biological evaluation of 1-{2,7-diazaspiro[3.5]nonan-2-yl}prop-2-en-1-one derivatives as covalent inhibitors of KRAS G12C with favorable metabolic stability and anti-tumor activity.
Bioorg.Med.Chem., 71, 2022
|
|
7YCB
 
 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE | | Descriptor: | CHLORIDE ION, GLYCEROL, Hydroxynitrile lyase, ... | | Authors: | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | Deposit date: | 2022-07-01 | | Release date: | 2024-01-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Based Site-Directed Mutagenesis of Hydroxynitrile Lyase from Cyanogenic Millipede, Oxidus gracilis for Hydrocyanation and Henry Reactions.
Chembiochem, 25, 2024
|
|
