1U96
 
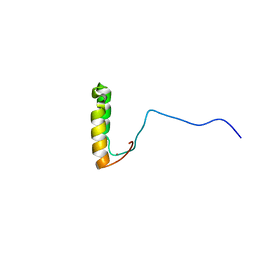 | | Solution Structure of Yeast Cox17 with Copper Bound | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase copper chaperone | | Authors: | Abajian, C, Yatsunyk, L.A, Ramirez, B.E, Rosenzweig, A.C. | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Yeast cox17 solution structure and Copper(I) binding.
J.Biol.Chem., 279, 2004
|
|
1U64
 
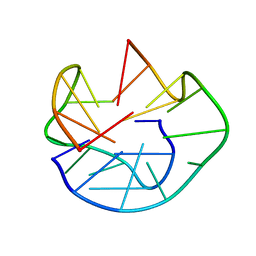 | | The Solution Structure of d(G3T4G4)2 | | Descriptor: | 5'-D(*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3' | | Authors: | Sket, P, Crnugelj, M, Plavec, J. | | Deposit date: | 2004-07-29 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | d(G3T4G4) forms unusual dimeric G-quadruplex structure with the same general fold in the presence of K+, Na+ or NH4+ ions.
Bioorg.Med.Chem., 12, 2004
|
|
1F84
 
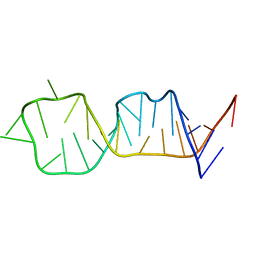 | | SOLUTION STRUCTURE OF HCV IRES RNA DOMAIN IIID | | Descriptor: | HCV-1B IRES RNA DOMAIN IIID | | Authors: | Lukavsky, P.J, Otto, G.A, Lancaster, A.M, Sarnow, P, Puglisi, J.D. | | Deposit date: | 2000-06-28 | | Release date: | 2000-11-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function.
Nat.Struct.Biol., 7, 2000
|
|
1HIT
 
 | |
1VMC
 
 | | STROMA CELL-DERIVED FACTOR-1ALPHA (SDF-1ALPHA) | | Descriptor: | Stromal cell-derived factor 1 | | Authors: | Gozansky, E.K, Clore, G.M. | | Deposit date: | 2004-09-20 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Mapping the binding of the N-terminal extracellular tail of the CXCR4 receptor to stromal cell-derived factor-1alpha.
J.Mol.Biol., 345, 2005
|
|
1HO6
 
 | |
1HRL
 
 | |
1HPN
 
 | | N.M.R. AND MOLECULAR-MODELLING STUDIES OF THE SOLUTION CONFORMATION OF HEPARIN | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose | | Authors: | Mulloy, B, Forster, M.J. | | Deposit date: | 1995-01-17 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | N.m.r. and molecular-modelling studies of the solution conformation of heparin.
Biochem.J., 293, 1993
|
|
2MQH
 
 | |
2JSN
 
 | |
2K3U
 
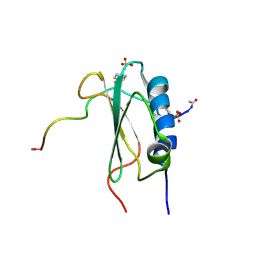 | | Structure of the tyrosine-sulfated C5a receptor N-terminus in complex with the immune evasion protein CHIPS. | | Descriptor: | C5a anaphylatoxin chemotactic receptor 1, Chemotaxis inhibitory protein | | Authors: | Ippel, J.H, Bunschoten, A, Kemmink, J, Liskamp, R. | | Deposit date: | 2008-05-16 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Tyrosine-sulfated C5a Receptor N Terminus in Complex with Chemotaxis Inhibitory Protein of Staphylococcus aureus.
J.Biol.Chem., 284, 2009
|
|
1JRW
 
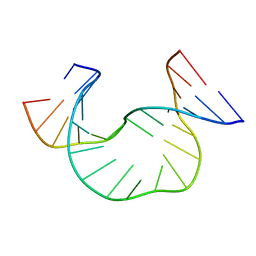 | | Solution Structure of dAATAA DNA Bulge | | Descriptor: | 5'-D(*CP*GP*TP*AP*GP*CP*CP*GP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*CP*GP*AP*AP*TP*AP*AP*GP*CP*TP*AP*CP*G)-3' | | Authors: | Gollmick, F.A, Lorenz, M, Dornberger, U, von Langen, J, Diekmann, S, Fritzsche, H. | | Deposit date: | 2001-08-15 | | Release date: | 2002-08-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dAATAA and dAAUAA DNA bulges.
Nucleic Acids Res., 30, 2002
|
|
1HME
 
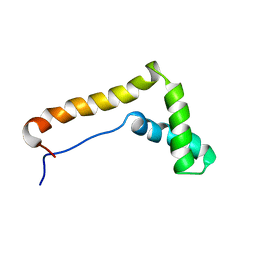 | | STRUCTURE OF THE HMG BOX MOTIF IN THE B-DOMAIN OF HMG1 | | Descriptor: | HIGH MOBILITY GROUP PROTEIN FRAGMENT-B | | Authors: | Weir, H.M, Kraulis, P.J, Hill, C.S, Raine, A.R.C, Laue, E.D, Thomas, J.O. | | Deposit date: | 1994-02-10 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the HMG box motif in the B-domain of HMG1.
EMBO J., 12, 1993
|
|
1JBF
 
 | |
1JI8
 
 | |
1JS5
 
 | | Solution Structure of dAAUAA DNA Bulge | | Descriptor: | 5'-D(*CP*GP*TP*AP*GP*CP*CP*GP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*CP*GP*AP*AP*UP*AP*AP*GP*CP*TP*AP*CP*G)-3' | | Authors: | Gollmick, F.A, Lorenz, M, Dornberger, U, von Langen, J, Diekmann, S, Fritzsche, H. | | Deposit date: | 2001-08-16 | | Release date: | 2002-08-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dAATAA and dAAUAA DNA bulges.
Nucleic Acids Res., 30, 2002
|
|
1Z0Q
 
 | | Aqueous Solution Structure of the Alzheimer's Disease Abeta Peptide (1-42) | | Descriptor: | Alzheimer's disease amyloid | | Authors: | Tomaselli, S, Esposito, V, Vangone, P, van Nuland, N.A, Bonvin, A.M, Guerrini, R, Tancredi, T, Temussi, P.A, Picone, D. | | Deposit date: | 2005-03-02 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The alpha-to-beta Conformational Transition of Alzheimer's Abeta-(1-42) Peptide in Aqueous Media is Reversible: A Step by Step Conformational Analysis Suggests the Location of beta Conformation Seeding
Chembiochem, 7, 2006
|
|
1JLO
 
 | |
1K1H
 
 | | HETERODUPLEX OF CHIRALLY PURE METHYLPHOSPHONATE/DNA DUPLEX | | Descriptor: | 5'-D(*CP*(CMR)P*(RMP)P*(RMP)P*(SMP)P*(CMR)P*(RMP))-3', 5'-D(*TP*GP*TP*TP*TP*GP*GP*C)-3' | | Authors: | Thiviyanathan, V, Vyazovkina, K.V, Gozansky, E.K, Bichenchova, E, Abramova, T.V, Luxon, B.A, Lebedev, A.V, Gorenstein, D.G. | | Deposit date: | 2001-09-25 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of hybrid backbone methylphosphonate DNA heteroduplexes: effect of R and S stereochemistry.
Biochemistry, 41, 2002
|
|
1KZV
 
 | |
1KZT
 
 | |
1IMU
 
 | | Solution Structure of HI0257, a Ribosome Binding Protein | | Descriptor: | HYPOTHETICAL PROTEIN HI0257 | | Authors: | Parsons, L, Eisenstein, E, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-11 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HI0257, a bacterial ribosome binding protein.
Biochemistry, 40, 2001
|
|
1KZS
 
 | |
1JTW
 
 | |
1JH3
 
 | | Solution structure of tyrosyl-tRNA synthetase C-terminal domain. | | Descriptor: | TYROSYL-TRNA SYNTHETASE | | Authors: | Guijarro, J.I, Pintar, A, Prochnicka-Chalufour, A, Guez, V, Gilquin, B, Bedouelle, H, Delepierre, M. | | Deposit date: | 2001-06-27 | | Release date: | 2002-03-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Anticodon Arm Binding Domain of Bacillus stearothermophilus
Tyrosyl-tRNA Synthetase
Structure, 10, 2002
|
|
