1F0E
 
 | | Cecropin A(1-8)-magainin 2(1-12) modified gig to P in dodecylphosphocholine micelles | | Descriptor: | CECROPIN A-MAGAININ 2 HYBRID PEPTIDE | | Authors: | Oh, D, Shin, S.Y, Lee, S, Kim, Y. | | Deposit date: | 2000-05-16 | | Release date: | 2000-06-14 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | Role of the hinge region and the tryptophan residue in the synthetic antimicrobial peptides, cecropin A(1-8)-magainin 2(1-12) and its analogues, on their antibiotic activities and structures.
Biochemistry, 39, 2000
|
|
1TXP
 
 | |
1TTX
 
 | | Solution Structure of human beta parvalbumin (oncomodulin) refined with a paramagnetism based strategy | | Descriptor: | CALCIUM ION, Oncomodulin | | Authors: | Babini, E, Bertini, I, Capozzi, F, Del Bianco, C, Hollender, D, Kiss, T, Luchinat, C, Quattrone, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-23 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human beta-Parvalbumin and Structural Comparison with Its Paralog alpha-Parvalbumin and with Their Rat Orthologs(,)
Biochemistry, 43, 2004
|
|
1U97
 
 | |
1DIU
 
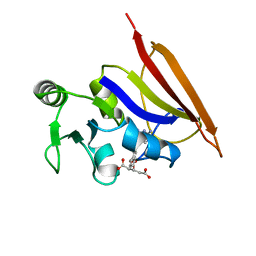 | | DIHYDROFOLATE REDUCTASE (E.C.1.5.1.3) COMPLEX WITH BRODIMOPRIM-4,6-DICARBOXYLATE | | Descriptor: | BRODIMOPRIM-4,6-DICARBOXYLATE, DIHYDROFOLATE REDUCTASE | | Authors: | Morgan, W.D, Birdsall, B, Polshakov, V.I, Sali, D, Kompis, I, Feeney, J. | | Deposit date: | 1995-08-01 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a brodimoprim analogue in its complex with Lactobacillus casei dihydrofolate reductase.
Biochemistry, 34, 1995
|
|
1DU9
 
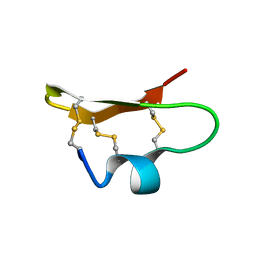 | | SOLUTION STRUCTURE OF BMP02, A NATURAL SCORPION TOXIN WHICH BLOCKS APAMIN-SENSITIVE CALCIUM-ACTIVATED POTASSIUM CHANNELS, 25 STRUCTURES | | Descriptor: | BMP02 NEUROTOXIN | | Authors: | Xu, Y, Wu, J, Pei, J, Shi, Y, Ji, Y, Tong, Q. | | Deposit date: | 2000-01-17 | | Release date: | 2000-02-04 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmP02, a new potassium channel blocker from the venom of the Chinese scorpion Buthus martensi Karsch.
Biochemistry, 39, 2000
|
|
1DX7
 
 | | Light-harvesting complex 1 beta subunit from Rhodobacter sphaeroides | | Descriptor: | Light harvesting 1 b(B850b) polypeptide | | Authors: | Conroy, M.J, Westerhuis, W, Parkes-Loach, P.S, Loach, P.A, Hunter, C.N, Williamson, M.P. | | Deposit date: | 1999-12-21 | | Release date: | 2000-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of Rhodobacter Sphaeroides Lh1 B Reveals Two Helical Domains Separated by a Flexible Region: Structural Consequences for the Lh1 Complex
J.Mol.Biol., 298, 2000
|
|
1SZY
 
 | |
1DSM
 
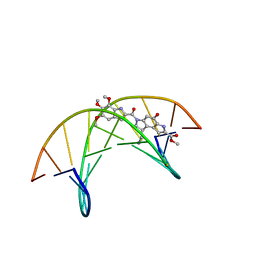 | | (-)-duocarmycin SA covalently linked to duplex DNA | | Descriptor: | 4-HYDROXY-8-METHYL-6-(4,5,6-TRIMETHOXY-1H-INDOLE-2-CARBONYL)-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, 5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3' | | Authors: | Smith, J.A, Case, D.A, Chazin, W.J. | | Deposit date: | 1999-03-27 | | Release date: | 1999-04-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The structural basis for in situ activation of DNA alkylation by duocarmycin SA
J.Mol.Biol., 300, 2000
|
|
1EXH
 
 | | SOLUTION STRUCTURE OF A CELLULOSE BINDING DOMAIN FROM CELLULOMONAS FIMI BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | EXO-1,4-BETA-D-GLYCANASE | | Authors: | Xu, G.-Y, Ong, E, Gilkes, N.R, Kilburn, D.G, Muhandiram, D.R, Harris-Brandts, M, Carver, J.P, Kay, L.E, Harvey, T.S. | | Deposit date: | 1995-03-14 | | Release date: | 1995-06-03 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
1F9L
 
 | |
1SG5
 
 | |
1FI9
 
 | | SOLUTION STRUCTURE OF THE IMIDAZOLE COMPLEX OF CYTOCHROME C | | Descriptor: | CYTOCHROME C, HEME C, IMIDAZOLE | | Authors: | Banci, L, Bertini, I, Liu, G, Lu, J, Reddig, T, Tang, W, Wu, Y, Zhu, D. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Effects of extrinsic imidazole ligation on the molecular and electronic structure of cytochrome c
J.Biol.Inorg.Chem., 6, 2001
|
|
1FBR
 
 | |
1FKT
 
 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | Descriptor: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | Authors: | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | Deposit date: | 1992-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1FKR
 
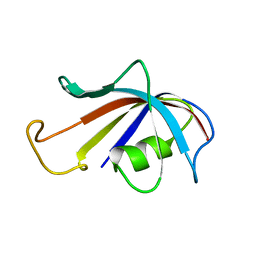 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | Descriptor: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | Authors: | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | Deposit date: | 1992-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1D70
 
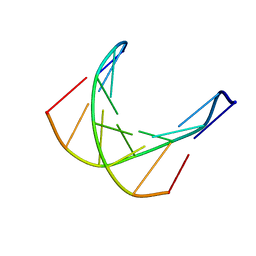 | |
1D2J
 
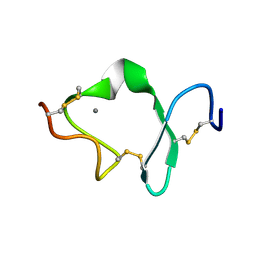 | | LDL RECEPTOR LIGAND-BINDING MODULE 6 | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR | | Authors: | North, C.L, Blacklow, S.C. | | Deposit date: | 1999-09-23 | | Release date: | 2000-03-22 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the sixth LDL-A module of the LDL receptor.
Biochemistry, 39, 2000
|
|
1FKS
 
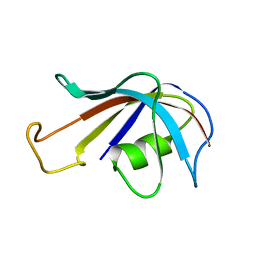 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | Descriptor: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | Authors: | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | Deposit date: | 1992-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1G84
 
 | | THE SOLUTION STRUCTURE OF THE C EPSILON2 DOMAIN FROM IGE | | Descriptor: | IMMUNOGLOBULIN E | | Authors: | McDonnell, J.M, Cowburn, D, Gould, H.J, Sutton, B.J, Calvert, R, Beavil, R.E, Beavil, A.J, Henry, A.J. | | Deposit date: | 2000-11-16 | | Release date: | 2001-05-16 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | The structure of the IgE Cepsilon2 domain and its role in stabilizing the complex with its high-affinity receptor FcepsilonRIalpha.
Nat.Struct.Biol., 8, 2001
|
|
1S7P
 
 | | Solution structure of thermolysin digested microcin J25 | | Descriptor: | microcin J25 | | Authors: | Rosengren, K.J, Blond, A, Afonso, C, Tabet, J.C, Rebuffat, S, Craik, D.J. | | Deposit date: | 2004-01-30 | | Release date: | 2004-06-15 | | Last modified: | 2011-07-27 | | Method: | SOLUTION NMR | | Cite: | Structure of thermolysin cleaved microcin J25: extreme stability of a two-chain antimicrobial peptide devoid of covalent links
Biochemistry, 43, 2004
|
|
1GJ2
 
 | | CO(III)-BLEOMYCIN-OOH BOUND TO AN OLIGONUCLEOTIDE CONTAINING A PHOSPHOGLYCOLATE LESION | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 5'-D(*CP*CP*AP*AP*AP*G)-3', 5'-D(*CP*CP*CP*AP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3', ... | | Authors: | Hoehn, S.T, Junker, H.-D, Bunt, R.C, Turner, C.J, Stubbe, J. | | Deposit date: | 2000-11-01 | | Release date: | 2001-06-06 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Co(III)-bleomycin-OOH bound to a phosphoglycolate lesion containing oligonucleotide: implications for bleomycin-induced double-strand DNA cleavage.
Biochemistry, 40, 2001
|
|
1GJG
 
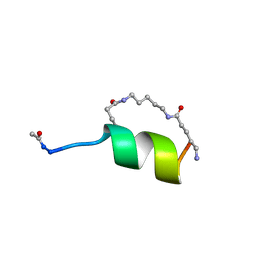 | | Peptide Antagonist of IGFBP1, (i,i+8) Covalently Restrained Analog, Minimized Average Structure | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1G5L
 
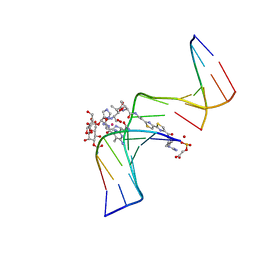 | | CO(III)-BLEOMYCIN-OOH BOUND TO AN OLIGONUCLEOTIDE CONTAINING A PHOSPHOGLYCOLATE LESION | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 5'-D(*CP*CP*AP*AP*AP*G)-3', 5'-D(*CP*CP*CP*AP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3', ... | | Authors: | Hoehn, S.T, Junker, H.-D, Bunt, R.C, Turner, C.J, Stubbe, J. | | Deposit date: | 2000-11-01 | | Release date: | 2001-06-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Co(III)-bleomycin-OOH bound to a phosphoglycolate lesion containing oligonucleotide: implications for bleomycin-induced double-strand DNA cleavage.
Biochemistry, 40, 2001
|
|
1SY8
 
 | | Structure of DNA sequence d-TGATCA by two-dimensional nuclear magnetic resonance spec and restrained molecular dynamics | | Descriptor: | 5'-D(P*TP*GP*AP*TP*CP*A)-3' | | Authors: | Barthwal, R, Awasthi, P, Narang, M, Sharma, U, Srivastava, N. | | Deposit date: | 2004-04-01 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of DNA sequence d-TGATCA by two-dimensional nuclear magnetic resonance spectroscopy and restrained molecular dynamics
J.STRUCT.BIOL., 148, 2004
|
|
