2N1Q
 
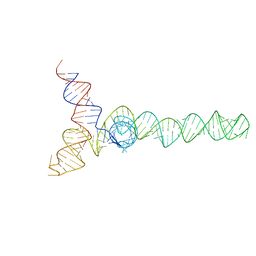 | | HIV-1 Core Packaging Signal | | Descriptor: | RNA_(155-MER) | | Authors: | Keane, S.C, Summers, M.F. | | Deposit date: | 2015-04-15 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RNA structure. Structure of the HIV-1 RNA packaging signal.
Science, 348, 2015
|
|
2OQ3
 
 | |
1Y9J
 
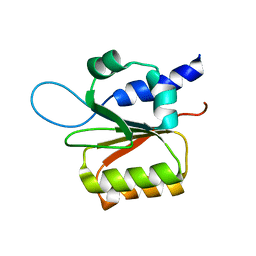 | | Solution structure of the rat Sly1 N-terminal domain | | Descriptor: | Sec1 family domain containing protein 1 | | Authors: | Arac, D, Dulubova, I, Pei, J, Huryeva, I, Grishin, N.V, Rizo, J. | | Deposit date: | 2004-12-15 | | Release date: | 2005-02-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional Structure of the rSly1 N-terminal Domain Reveals a Conformational Change Induced by Binding to Syntaxin 5.
J.Mol.Biol., 346, 2005
|
|
2MSW
 
 | |
2N9J
 
 | | Solution structure of oxidized human cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Imai, M, Saio, T, Kumeta, H, Uchida, T, Inagaki, F, Ishimori, K. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Investigation of the redox-dependent modulation of structure and dynamics in human cytochrome c.
Biochem.Biophys.Res.Commun., 469, 2016
|
|
2N9I
 
 | | Solution structure of reduced human cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Imai, M, Saio, T, Kumeta, H, Uchida, T, Inagaki, F, Ishimori, K. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Investigation of the redox-dependent modulation of structure and dynamics in human cytochrome c
Biochem.Biophys.Res.Commun., 469, 2016
|
|
1DIS
 
 | | DIHYDROFOLATE REDUCTASE (E.C.1.5.1.3) COMPLEX WITH BRODIMOPRIM-4,6-DICARBOXYLATE | | Descriptor: | BRODIMOPRIM-4,6-DICARBOXYLATE, DIHYDROFOLATE REDUCTASE | | Authors: | Morgan, W.D, Birdsall, B, Polshakov, V.I, Sali, D, Kompis, I, Feeney, J. | | Deposit date: | 1995-08-01 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a brodimoprim analogue in its complex with Lactobacillus casei dihydrofolate reductase.
Biochemistry, 34, 1995
|
|
1DB6
 
 | | SOLUTION STRUCTURE OF THE DNA APTAMER 5'-CGACCAACGTGTCGCCTGGTCG-3' COMPLEXED WITH ARGININAMIDE | | Descriptor: | ARGININEAMIDE, DNA | | Authors: | Robertson, S.A, Harada, K, Frankel, A.D, Wemmer, D.E. | | Deposit date: | 1999-11-02 | | Release date: | 2000-02-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure determination and binding kinetics of a DNA aptamer-argininamide complex.
Biochemistry, 39, 2000
|
|
1DBD
 
 | |
1EXL
 
 | | STRUCTURE OF AN 11-MER DNA DUPLEX CONTAINING THE CARBOCYCLIC NUCLEOTIDE ANALOG: 2'-DEOXYARISTEROMYCIN | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*GP*(2AR)P*GP*TP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*AP*CP*TP*CP*AP*CP*TP*G)-3') | | Authors: | Smirnov, S, Johnson, F, Marumoto, R, de los Santos, C. | | Deposit date: | 2000-05-03 | | Release date: | 2000-05-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an 11-mer DNA duplex containing the carbocyclic nucleotide analog: 2'-deoxyaristeromycin
J.Biomol.Struct.Dyn., 17, 2000
|
|
1EYO
 
 | |
1ESH
 
 | |
1UEO
 
 | | Solution structure of the [T8A]-Penaeidin-3 | | Descriptor: | Penaeidin-3a | | Authors: | Yang, Y, Poncet, J, Garnier, J, Zatylny, C, Bachere, E, Aumelas, A. | | Deposit date: | 2003-05-20 | | Release date: | 2003-10-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the recombinant penaeidin-3, a shrimp antimicrobial peptide
J.Biol.Chem., 278, 2003
|
|
1EW1
 
 | |
1TV0
 
 | | Solution structure of cryptdin-4, the most potent alpha-defensin from mouse Paneth cells | | Descriptor: | Cryptdin-4 | | Authors: | Jing, W, Hunter, H.N, Tanabe, H, Ouellette, A.J, Vogel, H.J. | | Deposit date: | 2004-06-25 | | Release date: | 2005-01-04 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cryptdin-4, a Mouse Paneth Cell alpha-Defensin.
Biochemistry, 43, 2004
|
|
1EXY
 
 | | SOLUTION STRUCTURE OF HTLV-1 PEPTIDE BOUND TO ITS RNA APTAMER TARGET | | Descriptor: | HTLV-1 REX PEPTIDE, RNA APTAMER, 33-MER | | Authors: | Jiang, F, Gorin, A, Hu, W, Majumdar, A, Baskerville, S, Xu, W, Ellington, A, Patel, D.J. | | Deposit date: | 2000-05-05 | | Release date: | 2000-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Anchoring an extended HTLV-1 Rex peptide within an RNA major groove containing junctional base triples.
Structure Fold.Des., 7, 1999
|
|
1EGL
 
 | |
1E4P
 
 | |
1F0D
 
 | | Cecropin A(1-8)-magainin 2(1-12) in dodecylphosphocholine micelles | | Descriptor: | CECROPIN A-MAGAININ 2 HYBRID PEPTIDE | | Authors: | Oh, D, Shin, S.Y, Lee, S, Kim, Y. | | Deposit date: | 2000-05-16 | | Release date: | 2000-06-14 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Role of the hinge region and the tryptophan residue in the synthetic antimicrobial peptides, cecropin A(1-8)-magainin 2(1-12) and its analogues, on their antibiotic activities and structures.
Biochemistry, 39, 2000
|
|
1F0F
 
 | | Cecropin A(1-8)-magainin 2(1-12) gig deletion modification in dodecylphosphocholine micelles | | Descriptor: | CECROPIN A-MAGAININ 2 HYBRID PEPTIDE | | Authors: | Oh, D, Shin, S.Y, Lee, S, Kim, Y. | | Deposit date: | 2000-05-16 | | Release date: | 2000-06-14 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Role of the hinge region and the tryptophan residue in the synthetic antimicrobial peptides, cecropin A(1-8)-magainin 2(1-12) and its analogues, on their antibiotic activities and structures.
Biochemistry, 39, 2000
|
|
1F0G
 
 | | Cecropin A(1-8)-magainin 2(1-12) L2 in dodecylphosphocholine micelles | | Descriptor: | CECROPIN A-MAGAININ 2 HYBRID PEPTIDE | | Authors: | Oh, D, Shin, S.Y, Lee, S, Kim, Y. | | Deposit date: | 2000-05-16 | | Release date: | 2000-06-14 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Role of the hinge region and the tryptophan residue in the synthetic antimicrobial peptides, cecropin A(1-8)-magainin 2(1-12) and its analogues, on their antibiotic activities and structures.
Biochemistry, 39, 2000
|
|
1F2H
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF THE TNFR1 ASSOCIATED PROTEIN, TRADD. | | Descriptor: | TUMOR NECROSIS FACTOR RECEPTOR TYPE 1 ASSOCIATED DEATH DOMAIN PROTEIN | | Authors: | Tsao, D, McDonaugh, T, Malakian, K, Xu, G.-Y, Telliez, J.-B, Hsu, H, Lin, L.-L. | | Deposit date: | 2000-05-24 | | Release date: | 2001-05-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of N-TRADD and characterization of the interaction of N-TRADD and C-TRAF2, a key step in the TNFR1 signaling pathway.
Mol.Cell, 5, 2000
|
|
1F0H
 
 | | Cecropin A(1-8)-magainin 2(1-12) A2 in dodecylphosphocholine micelles | | Descriptor: | CECROPIN A-MAGAININ 2 HYBRID PEPTIDE | | Authors: | Oh, D, Shin, S.Y, Lee, S, Kim, Y. | | Deposit date: | 2000-05-16 | | Release date: | 2000-06-14 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Role of the hinge region and the tryptophan residue in the synthetic antimicrobial peptides, cecropin A(1-8)-magainin 2(1-12) and its analogues, on their antibiotic activities and structures.
Biochemistry, 39, 2000
|
|
1RE6
 
 | | Localisation of Dynein Light Chains 1 and 2 and their Pro-apoptotic Ligands | | Descriptor: | dynein light chain 2 | | Authors: | Day, C.L, Puthalakath, H, Skea, G, Strasser, A, Barsukov, I, Lian, L.Y, Huang, D.C, Hinds, M.G. | | Deposit date: | 2003-11-06 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Localization of dynein light chains 1 and 2 and their pro-apoptotic ligands.
Biochem.J., 377, 2004
|
|
1EZE
 
 | | STRUCTURAL STUDIES OF A BABOON (PAPIO SP.) PLASMA PROTEIN INHIBITOR OF CHOLESTERYL ESTER TRANSFERASE. | | Descriptor: | CHOLESTERYL ESTER TRANSFERASE INHIBITOR PROTEIN | | Authors: | Buchko, G.W, Rozek, A, Kanda, P, Kennedy, M.A, Cushley, R.J. | | Deposit date: | 2000-05-10 | | Release date: | 2000-09-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural studies of a baboon (Papio sp.) plasma protein inhibitor of cholesteryl ester transferase.
Protein Sci., 9, 2000
|
|
