5BWM
 
 | | The complex structure of C3cer exoenzyme and GDP bound RhoA (NADH-bound state) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADP-ribosyltransferase, ... | | Authors: | Toda, A, Tsurumura, T, Yoshida, T, Tsuge, H. | | Deposit date: | 2015-06-08 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rho GTPase Recognition by C3 Exoenzyme Based on C3-RhoA Complex Structure.
J.Biol.Chem., 290, 2015
|
|
2A78
 
 | | Crystal structure of the C3bot-RalA complex reveals a novel type of action of a bacterial exoenzyme | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mono-ADP-ribosyltransferase C3, ... | | Authors: | Pautsch, A, Vogelsgesang, M, Trankle, J, Herrmann, C, Aktories, K. | | Deposit date: | 2005-07-05 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of the C3bot-RalA complex reveals a novel type of action of a bacterial exoenzyme.
Embo J., 24, 2005
|
|
7YVS
 
 | | Complex structure of Clostridioides difficile binary toxin unfolded CDTa-bound CDTb-pore (short). | | Descriptor: | ADP-ribosylating binary toxin binding subunit CdtB, ADP-ribosylating binary toxin enzymatic subunit CdtA, CALCIUM ION | | Authors: | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | Deposit date: | 2022-08-19 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7YVQ
 
 | | Complex structure of Clostridioides difficile binary toxin folded CDTa-bound CDTb-pore (short). | | Descriptor: | ADP-ribosylating binary toxin binding subunit CdtB, ADP-ribosylating binary toxin enzymatic subunit CdtA, CALCIUM ION | | Authors: | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | Deposit date: | 2022-08-19 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7VNN
 
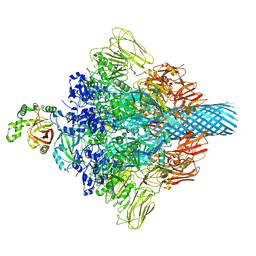 | | Complex structure of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) pore with long stem | | Descriptor: | ADP-ribosylating binary toxin binding subunit CdtB, CALCIUM ION, CdtA | | Authors: | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | Deposit date: | 2021-10-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7VNJ
 
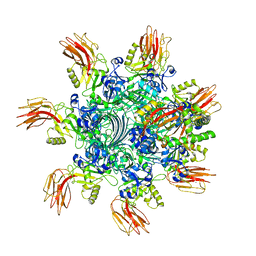 | | Complex structure of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) pore with short stem | | Descriptor: | ADP-ribosylating binary toxin binding subunit CdtB, ADP-ribosyltransferase enzymatic component, CALCIUM ION | | Authors: | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | Deposit date: | 2021-10-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
3BUZ
 
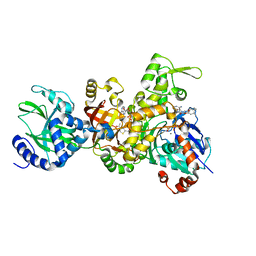 | | Crystal structure of ia-bTAD-actin complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Tsuge, H, Nagahama, M, Oda, M, Iwamoto, S, Utsunomiya, H, Marquez, V.E, Katunuma, N, Nishizawa, M, Sakurai, J. | | Deposit date: | 2008-01-04 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis of actin recognition and arginine ADP-ribosylation by Clostridium perfringens iota-toxin
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2A9K
 
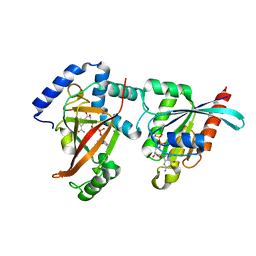 | | Crystal structure of the C3bot-NAD-RalA complex reveals a novel type of action of a bacterial exoenzyme | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mono-ADP-ribosyltransferase C3, ... | | Authors: | Pautsch, A, Vogelsgesang, M, Trankle, J, Herrmann, C, Aktories, K. | | Deposit date: | 2005-07-12 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of the C3bot-RalA complex reveals a novel type of action of a bacterial exoenzyme.
Embo J., 24, 2005
|
|
6GNK
 
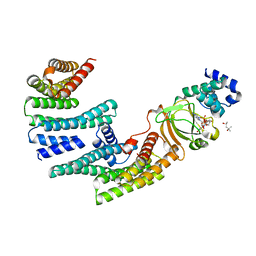 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, trimeric crystal form bound to Carba-NAD | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 14-3-3 protein beta/alpha, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
6GNJ
 
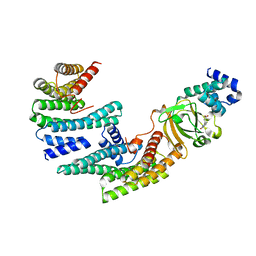 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, trimeric crystal form in complex with STO1101 | | Descriptor: | 14-3-3 protein beta/alpha, 3-(12-oxidanylidene-7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(8),2(6),9-trien-10-yl)propanoic acid, Exoenzyme S | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
6GN0
 
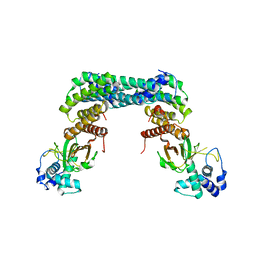 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, tetrameric crystal form | | Descriptor: | 14-3-3 protein beta/alpha, Exoenzyme S | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-29 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
6GNN
 
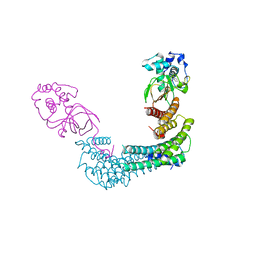 | | Exoenzyme T from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, tetrameric crystal form bound to STO1101 | | Descriptor: | 14-3-3 protein beta/alpha, 3-(12-oxidanylidene-7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(8),2(6),9-trien-10-yl)propanoic acid, Exoenzyme T | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
6GN8
 
 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, trimeric crystal form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 14-3-3 protein beta/alpha, Exoenzyme S | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
4XSH
 
 | | The complex structure of C3cer exoenzyme and GTP bound RhoA (NADH-bound state) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Toda, A, Tsurumura, T, Yoshida, T, Tsuge, H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rho GTPase Recognition by C3 Exoenzyme Based on C3-RhoA Complex Structure.
J.Biol.Chem., 290, 2015
|
|
4XSG
 
 | | The complex structure of C3cer exoenzyme and GTP bound RhoA (NADH-free state) | | Descriptor: | 1,2-ETHANEDIOL, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ADP-ribosyltransferase, ... | | Authors: | Toda, A, Tsurumura, T, Yoshida, T, Tsuge, H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rho GTPase Recognition by C3 Exoenzyme Based on C3-RhoA Complex Structure.
J.Biol.Chem., 290, 2015
|
|
4GY2
 
 | | Crystal structure of apo-Ia-actin complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Tsurumura, T, Oda, M, Nagahama, M, Tsuge, H. | | Deposit date: | 2012-09-05 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Arginine ADP-ribosylation mechanism based on structural snapshots of iota-toxin and actin complex
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4H0X
 
 | | Crystal structure of NAD+-Ia(E380A)-actin complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Tsurumura, T, Oda, M, Nagahama, M, Tsuge, H. | | Deposit date: | 2012-09-10 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Arginine ADP-ribosylation mechanism based on structural snapshots of iota-toxin and actin complex
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4H0T
 
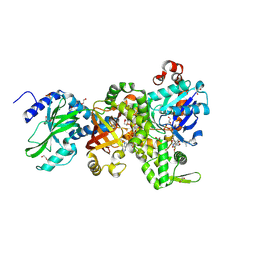 | | Crystal structure of Ia-ADPR-actin complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Tsurumura, T, Oda, M, Nagahama, M, Tsuge, H. | | Deposit date: | 2012-09-10 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arginine ADP-ribosylation mechanism based on structural snapshots of iota-toxin and actin complex
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4H0Y
 
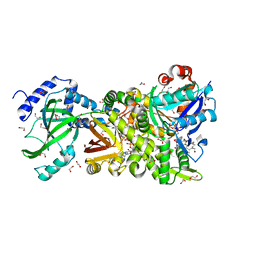 | | Crystal structure of NAD+-Ia(E380S)-actin complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Tsurumura, T, Oda, M, Nagahama, M, Tsuge, H. | | Deposit date: | 2012-09-10 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Arginine ADP-ribosylation mechanism based on structural snapshots of iota-toxin and actin complex
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4H0V
 
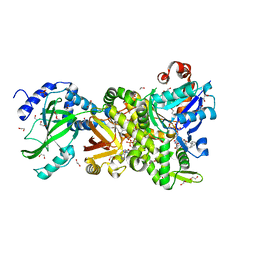 | | Crystal structure of NAD+-Ia(E378S)-actin complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Tsurumura, T, Oda, M, Nagahama, M, Tsuge, H. | | Deposit date: | 2012-09-10 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Arginine ADP-ribosylation mechanism based on structural snapshots of iota-toxin and actin complex
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4H03
 
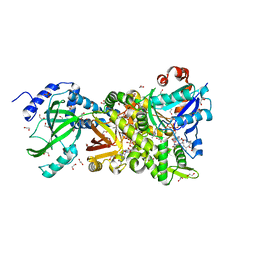 | | Crystal structure of NAD+-Ia-actin complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Tsurumura, T, Oda, M, Nagahama, M, Tsuge, H. | | Deposit date: | 2012-09-07 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Arginine ADP-ribosylation mechanism based on structural snapshots of iota-toxin and actin complex
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
