6HPW
 
 | | Crystal structure of ENL (MLLT1) in complex with compound 20 | | Descriptor: | 1,2-ETHANEDIOL, 3-iodanyl-4-methyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
6HQ0
 
 | | Crystal structure of ENL (MLLT1), apo form | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
6HT0
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with compound 94 | | Descriptor: | 1,2-ETHANEDIOL, 1-cyclopropyl-~{N}-[2-[[(2~{S})-2-methylpyrrolidin-1-yl]methyl]-3~{H}-benzimidazol-5-yl]indazole-5-carboxamide, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-10-02 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of an MLLT1/3 YEATS Domain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7E74
 
 | |
7E7A
 
 | | Crystal structure of apo ENL YEATS domain T3 mutant | | Descriptor: | Protein ENL | | Authors: | Li, Y, Li, H. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of ENL YEATS domain T1 mutant in complex with histone H3 acetylation at K27
To Be Published
|
|
7EIC
 
 | |
7EID
 
 | |
7EIF
 
 | | Crystal structure of GAS41 YEATS domain | | Descriptor: | GLYCEROL, YEATS domain-containing protein 4 | | Authors: | Kikuchi, M, Umehara, T. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | GAS41 promotes H2A.Z deposition through recognition of the N terminus of histone H3 by the YEATS domain.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7E7C
 
 | |
7EIE
 
 | | Crystal structure of YEATS2 YEATS domain | | Descriptor: | GLYCEROL, YEATS domain-containing protein 2 | | Authors: | Kikuchi, M, Umehara, T. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Elucidation of binding preferences of YEATS domains to site-specific acetylated nucleosome core particles.
J.Biol.Chem., 298, 2022
|
|
6L5Z
 
 | |
6MIN
 
 | |
6MIM
 
 | |
6MIO
 
 | | Crystal structure of Taf14 YEATS domain in complex with histone H3K9pr | | Descriptor: | Histone H3K9pr, Transcription initiation factor TFIID subunit 14 | | Authors: | Klein, B.J, Andrews, F.H, Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2018-09-19 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
6MIQ
 
 | | Crystal structure of Taf14 YEATS domain in complex with histone H3K9bu | | Descriptor: | Histone H3K9bu, Transcription initiation factor TFIID subunit 14 | | Authors: | Klein, B.J, Andrews, F.H, Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2018-09-19 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
6MIL
 
 | | Crystal structure of AF9 YEATS domain in complex with histone H3K9bu | | Descriptor: | DI(HYDROXYETHYL)ETHER, Histone H3K9bu, MALONATE ION, ... | | Authors: | Vann, K.R, Klein, B.J, Kutateladze, T.G. | | Deposit date: | 2018-09-19 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
6LS6
 
 | |
6LSD
 
 | |
6MIP
 
 | |
5SVA
 
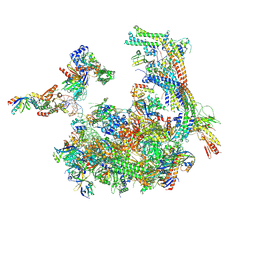 | | Mediator-RNA Polymerase II Pre-Initiation Complex | | Descriptor: | 108bp HIS4 Promoter Non-template Strand (-92/+16), 108bp HIS4 Promoter Template Strand (+16/-92), DNA repair helicase RAD25, ... | | Authors: | Robinson, P.J, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2016-08-05 | | Release date: | 2016-09-28 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (15.3 Å) | | Cite: | Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex.
Cell, 166, 2016
|
|
8X15
 
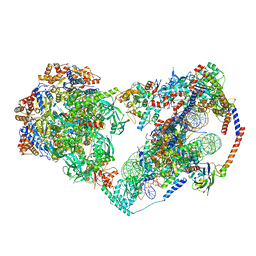 | | Structure of nucleosome-bound SRCAP-C in the apo state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
8X1C
 
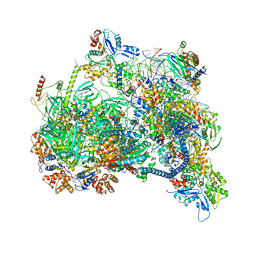 | | Structure of nucleosome-bound SRCAP-C in the ADP-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
8X19
 
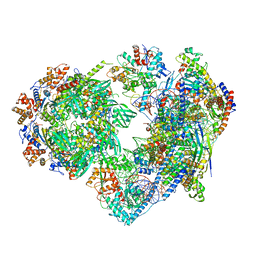 | | Structure of nucleosome-bound SRCAP-C in the ADP-BeFx-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
8IIY
 
 | |
8IIZ
 
 | |
