3ZLH
 
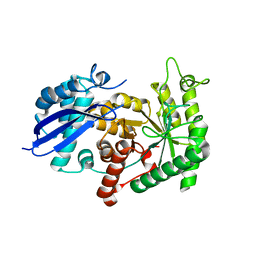 | | Structure of group A Streptococcal enolase | | Descriptor: | ENOLASE | | Authors: | Cork, A.J, Ericsson, D.J, Law, R.H.P, Casey, L.W, Valkov, E, Bertozzi, C, Stamp, A, Aquilina, J.A, Whisstock, J.C, Walker, M.J, Kobe, B. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Stability of the Octameric Structure Affects Plasminogen-Binding Capacity of Streptococcal Enolase.
Plos One, 10, 2015
|
|
3ZLF
 
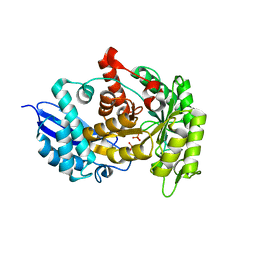 | | Structure of group A Streptococcal enolase K312A mutant | | Descriptor: | ENOLASE, PHOSPHATE ION | | Authors: | Cork, A.J, Ericsson, D.J, Law, R.H.P, Casey, L.W, Valkov, E, Bertozzi, C, Stamp, A, Aquilina, J.A, Whisstock, J.C, Walker, M.J, Kobe, B. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Stability of the Octameric Structure Affects Plasminogen-Binding Capacity of Streptococcal Enolase.
Plos One, 10, 2015
|
|
4EWJ
 
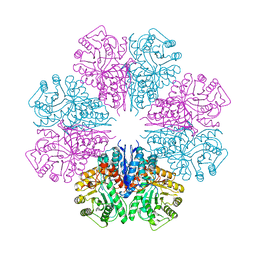 | | structure of the enloase from Streptococcus suis serotype 2 | | Descriptor: | Enolase 2 | | Authors: | Lu, Q, Lu, H, Qi, J, Lu, G, Gao, G.F. | | Deposit date: | 2012-04-27 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | An octamer of enolase from Streptococcus suis.
Protein Cell, 3, 2012
|
|
4G7F
 
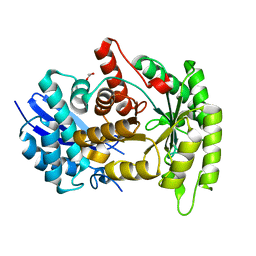 | |
3UCC
 
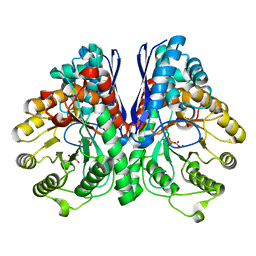 | | Asymmetric complex of human neuron specific enolase-1-PGA/PEP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-PHOSPHOGLYCERIC ACID, Gamma-enolase, ... | | Authors: | Qin, J, Chai, G, Brewer, J, Lovelace, L, Lebioda, L. | | Deposit date: | 2011-10-26 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity.
J.Inorg.Biochem., 111, 2012
|
|
3UJF
 
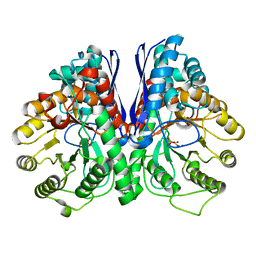 | | Asymmetric complex of human neuron specific enolase-4-PGA/PEP | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, Gamma-enolase, MAGNESIUM ION, ... | | Authors: | Qin, J, Chai, G, Brewer, J, Lovelace, L, Lebioda, L. | | Deposit date: | 2011-11-07 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity.
J.Inorg.Biochem., 111, 2012
|
|
3UJR
 
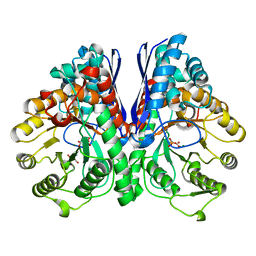 | | Asymmetric complex of human neuron specific enolase-5-PGA/PEP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-PHOSPHOGLYCERIC ACID, Gamma-enolase, ... | | Authors: | Qin, J, Chai, G, Brewer, J, Lovelace, L, Lebioda, L. | | Deposit date: | 2011-11-08 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity.
J.Inorg.Biochem., 111, 2012
|
|
3UCD
 
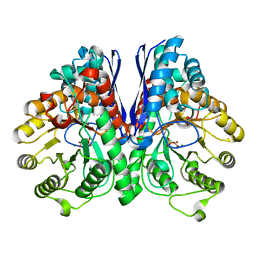 | | Asymmetric complex of human neuron specific enolase-2-PGA/PEP | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, Gamma-enolase, MAGNESIUM ION, ... | | Authors: | Qin, J, Chai, G, Brewer, J, Lovelace, L, Lebioda, L. | | Deposit date: | 2011-10-26 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity.
J.Inorg.Biochem., 111, 2012
|
|
3UJE
 
 | | Asymmetric complex of human neuron specific enolase-3-PGA/PEP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-PHOSPHOGLYCERIC ACID, Gamma-enolase, ... | | Authors: | Qin, J, Chai, G, Brewer, J, Lovelace, L, Lebioda, L. | | Deposit date: | 2011-11-07 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity.
J.Inorg.Biochem., 111, 2012
|
|
3UJS
 
 | | Asymmetric complex of human neuron specific enolase-6-PGA/PEP | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, (2R)-3-oxo-2-(phosphonooxy)propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Qin, J, Chai, G, Brewer, J, Lovelace, L, Lebioda, L. | | Deposit date: | 2011-11-08 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity.
J.Inorg.Biochem., 111, 2012
|
|
4A3R
 
 | | Crystal structure of Enolase from Bacillus subtilis. | | Descriptor: | CITRIC ACID, ENOLASE, SODIUM ION | | Authors: | Newman, J.A, Hewitt, L, Rodrigues, C, Solovyova, A.S, Harwood, C.R, Lewis, R.J. | | Deposit date: | 2011-10-04 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dissection of the Network of Interactions that Links RNA Processing with Glycolysis in the Bacillus Subtilis Degradosome.
J.Mol.Biol., 416, 2012
|
|
3UJ2
 
 | | CRYSTAL STRUCTURE OF AN ENOLASE FROM ANAEROSTIPES CACCAE (EFI TARGET EFI-502054) WITH BOUND MG AND SULFATE | | Descriptor: | Enolase 1, MAGNESIUM ION, SULFATE ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ENOLASE FROM ANAEROSTIPES CACCAE (EFI TARGET EFI-502054) WITH BOUND MG AND SULFATE
to be published
|
|
3TQP
 
 | | Structure of an enolase (eno) from Coxiella burnetii | | Descriptor: | Enolase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Rudolph, M, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3QTP
 
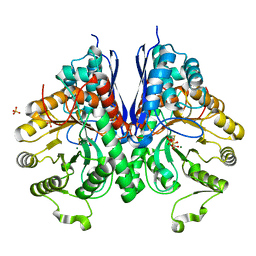 | |
3QN3
 
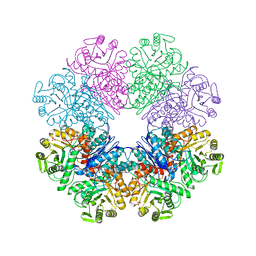 | | Phosphopyruvate hydratase from Campylobacter jejuni. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Enolase, GLYCEROL, ... | | Authors: | Osipiuk, J, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Phosphopyruvate hydratase from Campylobacter jejuni.
To be Published
|
|
2XSX
 
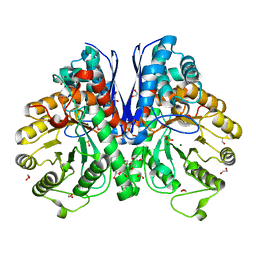 | | Crystal structure of human beta enolase ENOB | | Descriptor: | 1,2-ETHANEDIOL, BETA-ENOLASE, MAGNESIUM ION, ... | | Authors: | Vollmar, M, Krysztofinska, E, Chaikuad, A, Krojer, T, Cocking, R, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Yue, W.W, Oppermann, U. | | Deposit date: | 2010-09-30 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Human Beta Enolase Enob
To be Published
|
|
3OTR
 
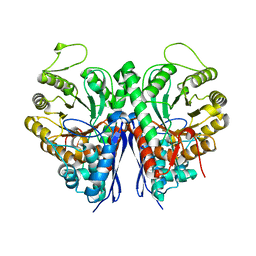 | | 2.75 Angstrom Crystal Structure of Enolase 1 from Toxoplasma gondii | | Descriptor: | CHLORIDE ION, Enolase, SULFATE ION | | Authors: | Minasov, G, Ruan, J, Shuvalova, L, Halavaty, A, Ngo, H, Tomavo, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-13 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure of bradyzoite-specific enolase from Toxoplasma gondii reveals insights into its dual cytoplasmic and nuclear functions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2XH2
 
 | |
2XGZ
 
 | |
2XH4
 
 | |
2XH7
 
 | |
2XH0
 
 | |
3H8A
 
 | |
3B97
 
 | | Crystal Structure of human Enolase 1 | | Descriptor: | Alpha-enolase, MAGNESIUM ION, SULFATE ION | | Authors: | Kang, H.J, Jung, S.K, Kim, S.J, Chung, S.J. | | Deposit date: | 2007-11-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human alpha-enolase (hENO1), a multifunctional glycolytic enzyme.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2PSN
 
 | |
