4PP4
 
 | |
1R5N
 
 | | Crystal Structure Analysis of sup35 complexed with GDP | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Kong, C, Song, H. | | Deposit date: | 2003-10-10 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe
Mol.Cell, 14, 2004
|
|
4JZZ
 
 | |
4JZW
 
 | |
4R4N
 
 | | Crystal structure of the anti-hiv-1 antibody 2.2c in complex with hiv-1 93ug037 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2.2c LIGHT CHAIN, Antibody 2.2c heavy CHAIN, ... | | Authors: | Acharya, P, Louder, R, Kwong, P.D. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
4K0A
 
 | |
3R26
 
 | | Perrhenate Binding to Molybdate Binding Protein | | Descriptor: | Molybdate-binding periplasmic protein, PERRHENATE | | Authors: | Aryal, B.P, Brugarolas, P, He, C. | | Deposit date: | 2011-03-13 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of ReO(4) (-) with an engineered MoO (4) (2-)-binding protein: towards a new approach in radiopharmaceutical applications.
J.Biol.Inorg.Chem., 17, 2012
|
|
3US4
 
 | |
4U6W
 
 | |
4U71
 
 | |
4U75
 
 | | HsMetAP (F309M) in complex with Methionine | | Descriptor: | COBALT (II) ION, GLYCEROL, METHIONINE, ... | | Authors: | Arya, T, Addlagatta, A. | | Deposit date: | 2014-07-30 | | Release date: | 2015-03-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification of the Molecular Basis of Inhibitor Selectivity between the Human and Streptococcal Type I Methionine Aminopeptidases
J.Med.Chem., 58, 2015
|
|
3PDM
 
 | | Hibiscus Latent Singapore virus | | Descriptor: | Coat protein, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Tewary, S.K, Wong, S.M, Swaminathan, K. | | Deposit date: | 2010-10-22 | | Release date: | 2011-01-12 | | Last modified: | 2024-03-20 | | Method: | FIBER DIFFRACTION (3.5 Å) | | Cite: | Structure of Hibiscus latent Singapore virus by fiber diffraction: A non-conserved His122 contributes to coat protein stability
J.Mol.Biol., 2010
|
|
4KW3
 
 | |
3T4U
 
 | |
1GK1
 
 | | Structure-based prediction of modifications in glutarylamidase to allow single-step enzymatic production of 7-aminocephalosporanic acid from cephalosporin C | | Descriptor: | CEPHALOSPORIN ACYLASE, GLYCEROL | | Authors: | Fritz-Wolf, K, Koller, K.P, Lange, G, Liesum, A, Sauber, K, Schreuder, H, Aretz, W, Kabsch, W. | | Deposit date: | 2001-08-07 | | Release date: | 2002-01-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Prediction of Modifications in Glutarylamidase to Allow Single-Step Enzymatic Production of 7-Aminocephalosporanic Acid from Cephalosporin C.
Protein Sci., 11, 2002
|
|
1GK0
 
 | | Structure-based prediction of modifications in glutarylamidase to allow single-step enzymatic production of 7-aminocephalosporanic acid from cephalosporin C | | Descriptor: | 1,2-ETHANEDIOL, CEPHALOSPORIN ACYLASE, PHOSPHATE ION | | Authors: | Fritz-Wolf, K, Koller, K.P, Lange, G, Liesum, A, Sauber, K, Schreuder, H, Aretz, W, Kabsch, W. | | Deposit date: | 2001-08-07 | | Release date: | 2002-01-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Prediction of Modifications in Glutarylamidase to Allow Single-Step Enzymatic Production of 7-Aminocephalosporanic Acid from Cephalosporin C.
Protein Sci., 11, 2002
|
|
5TI1
 
 | |
3WJM
 
 | | Crystal structure of Bombyx mori Sp2/Sp3 heterohexamer | | Descriptor: | Arylphorin, Silkworm storage protein, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yuan, Y.A, Hou, Y. | | Deposit date: | 2013-10-11 | | Release date: | 2014-09-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Bombyx mori arylphorins reveals a 3:3 heterohexamer with multiple papain cleavage sites
Protein Sci., 23, 2014
|
|
3VU0
 
 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (AfAglB-S2, AF_0040, O30195_ARCFU) from Archaeoglobus fulgidus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative uncharacterized protein | | Authors: | Nyirenda, J, Matsumoto, S, Saitoh, T, Maita, N, Noda, N.N, Inagaki, F, Kohda, D. | | Deposit date: | 2012-06-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystallographic and NMR Evidence for Flexibility in Oligosaccharyltransferases and Its Catalytic Significance
Structure, 21, 2013
|
|
3VU1
 
 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (PhAglB-L, O74088_PYRHO) from Pyrococcus horikoshii | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative uncharacterized protein PH0242 | | Authors: | Nyirenda, J, Matsumoto, S, Saitoh, T, Maita, N, Noda, N.N, Inagaki, F, Kohda, D. | | Deposit date: | 2012-06-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic and NMR Evidence for Flexibility in Oligosaccharyltransferases and Its Catalytic Significance
Structure, 21, 2013
|
|
1ILV
 
 | | Crystal Structure Analysis of the TM107 | | Descriptor: | STATIONARY-PHASE SURVIVAL PROTEIN SURE HOMOLOG | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Beasley, S, Evdokimova, E, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-05-08 | | Release date: | 2001-10-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase.
Structure, 9, 2001
|
|
2E9H
 
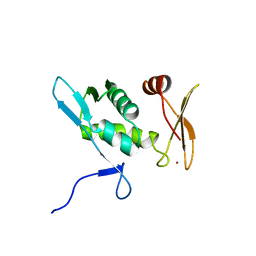 | | Solution structure of the eIF-5_eIF-2B domain from human Eukaryotic translation initiation factor 5 | | Descriptor: | Eukaryotic translation initiation factor 5, ZINC ION | | Authors: | Tomizawa, T, Tochio, N, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-25 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the eIF-5_eIF-2B domain from human Eukaryotic translation initiation factor 5
To be Published
|
|
4AXP
 
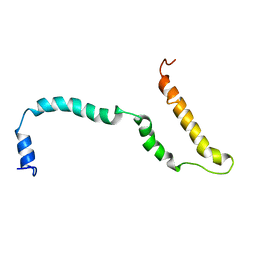 | | NMR structure of Hsp12, a protein induced by and required for dietary restriction-induced lifespan extension in yeast. | | Descriptor: | 12 KDA HEAT SHOCK PROTEIN | | Authors: | Herbert, A.P, Riesen, M, Bloxam, L, Kosmidou, E, Wareing, B.M, Johnson, J.R, Phelan, M.M, Pennington, S.R, Lian, L.Y, Morgan, A. | | Deposit date: | 2012-06-13 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Hsp12, a Protein Induced by and Required for Dietary Restriction-Induced Lifespan Extension in Yeast.
Plos One, 7, 2012
|
|
1XTD
 
 | |
1FRA
 
 | |
