1KNA
 
 | |
1KNE
 
 | |
1JGG
 
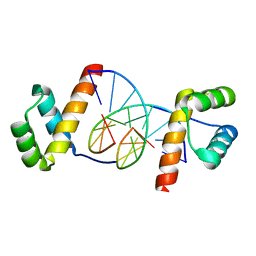 | | Even-skipped Homeodomain Complexed to AT-rich DNA | | Descriptor: | 5'-D(P*AP*AP*TP*TP*CP*AP*AP*TP*TP*A)-3', 5'-D(P*TP*AP*AP*TP*TP*GP*AP*AP*TP*T)-3', Segmentation Protein Even-Skipped | | Authors: | Hirsch, J.A, Aggarwal, A.K. | | Deposit date: | 2001-06-25 | | Release date: | 2001-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the even-skipped homeodomain complexed to AT-rich DNA: new perspectives on homeodomain specificity.
EMBO J., 14, 1995
|
|
5ONB
 
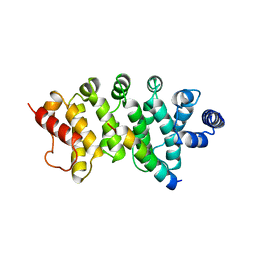 | | Drosophila Bag-of-marbles CBM peptide bound to human CAF40 | | Descriptor: | CCR4-NOT transcription complex subunit 9, GLYCEROL, Protein bag-of-marbles | | Authors: | Raisch, T, Sgromo, A, Backhaus, C, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2017-08-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DrosophilaBag-of-marbles directly interacts with the CAF40 subunit of the CCR4-NOT complex to elicit repression of mRNA targets.
RNA, 24, 2018
|
|
1KW4
 
 | |
1MXE
 
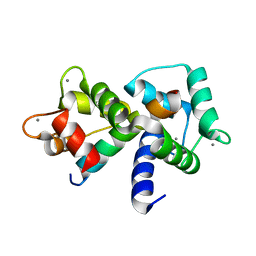 | | Structure of the Complex of Calmodulin with the Target Sequence of CaMKI | | Descriptor: | CALCIUM ION, Calmodulin, Target Sequence of rat Calmodulin-Dependent Protein Kinase I | | Authors: | Clapperton, J.A, Martin, S.R, Smerdon, S.J, Gamblin, S.J, Bayley, P.M. | | Deposit date: | 2002-10-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Complex
of Calmodulin with the Target
Sequence of Calmodulin-Dependent
Protein Kinase I: Studies of the
Kinase Activation Mechanism
Biochemistry, 41, 2002
|
|
1N6M
 
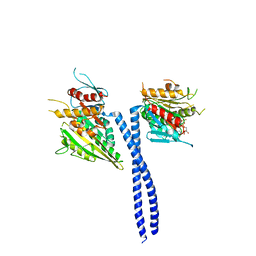 | | Rotation of the stalk/neck and one head in a new crystal structure of the kinesin motor protein, Ncd | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Claret segregational protein, MAGNESIUM ION | | Authors: | Yun, M, Bronner, C.E, Park, C.-G, Cha, S.-S, Park, H.-W, Endow, S.A. | | Deposit date: | 2002-11-11 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rotation of the stalk/neck and one head in a new crystal structure of the kinesin motor protein, Ncd
EMBO J., 22, 2003
|
|
3BRL
 
 | |
3A0B
 
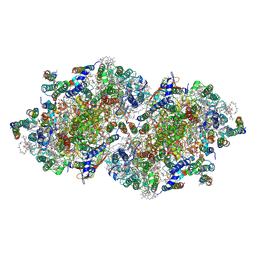 | | Crystal structure of Br-substituted Photosystem II complex | | Descriptor: | (1S)-2-(ALPHA-L-ALLOPYRANOSYLOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYL PALMITATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 5-[(2E,6E,10E,14E,18E,22E)-3,7,11,15,19,23,27-HEPTAMETHYLOCTACOSA-2,6,10,14,18,22,26-HEPTAENYL]-2,3-DIMETHYLBENZO-1,4-QUINONE, ... | | Authors: | Kawakami, K, Umena, Y, Kamiya, N, Shen, J.-R. | | Deposit date: | 2009-03-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Location of chloride and its possible functions in oxygen-evolving photosystem II revealed by X-ray crystallography
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3A0H
 
 | | Crystal structure of I-substituted Photosystem II complex | | Descriptor: | (1S)-2-(ALPHA-L-ALLOPYRANOSYLOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYL PALMITATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 5-[(2E,6E,10E,14E,18E,22E)-3,7,11,15,19,23,27-HEPTAMETHYLOCTACOSA-2,6,10,14,18,22,26-HEPTAENYL]-2,3-DIMETHYLBENZO-1,4-QUINONE, ... | | Authors: | Kawakami, K, Umena, Y, Kamiya, N, Shen, J.-R. | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Location of chloride and its possible functions in oxygen-evolving photosystem II revealed by X-ray crystallography
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1OO0
 
 | |
2J59
 
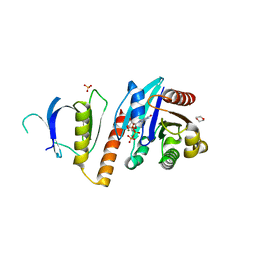 | | Crystal structure of the ARF1:ARHGAP21-ArfBD complex | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, ADP-RIBOSYLATION FACTOR 1, ... | | Authors: | Menetrey, J, Perderiset, M, Cicolari, J, Dubois, T, El Khatib, N, El Khadali, F, Franco, M, Chavrier, P, Houdusse, A. | | Deposit date: | 2006-09-13 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Arf1-Mediated Recruitment of Arhgap21 to Golgi Membranes.
Embo J., 26, 2007
|
|
1OT3
 
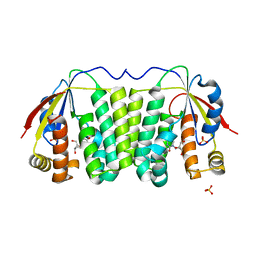 | | Crystal structure of Drosophila deoxyribonucleotide kinase complexed with the substrate deoxythymidine | | Descriptor: | Deoxyribonucleoside Kinase, SULFATE ION, THYMIDINE | | Authors: | Mikkelsen, N.E, Johansson, K, Karlsson, A, Knecht, W, Andersen, G, Piskur, J, Munch-Petersen, B, Eklund, H. | | Deposit date: | 2003-03-21 | | Release date: | 2003-05-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Feedback Inhibition of the Deoxyribonucleoside Salvage Pathway: Studies of the Drosophila Deoxyribonucleoside Kinase.
Biochemistry, 42, 2003
|
|
1PDU
 
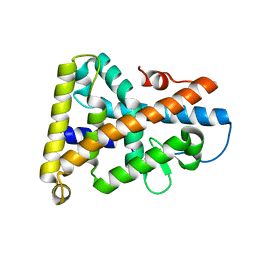 | | Ligand-binding domain of Drosophila orphan nuclear receptor DHR38 | | Descriptor: | nuclear hormone receptor HR38 | | Authors: | Baker, K.D, Shewchuk, L.M, Korlova, T, Makishima, M, Hassell, A.M, Wisely, B, Caravella, J.A, Lambert, M.H, Wilson, T.M, Mangelsdorf, D.J. | | Deposit date: | 2003-05-20 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway.
Cell(Cambridge,Mass.), 113, 2003
|
|
1PDQ
 
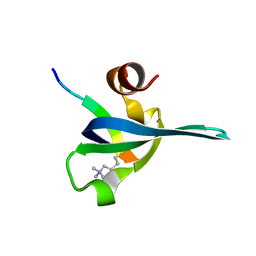 | | Polycomb chromodomain complexed with the histone H3 tail containing trimethyllysine 27. | | Descriptor: | Histone H3.3, Polycomb protein | | Authors: | Fischle, W, Wang, Y, Jacobs, S.A, Kim, Y, Allis, C.D, Khorasanizadeh, S. | | Deposit date: | 2003-05-20 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains
Genes Dev., 17, 2003
|
|
1PDN
 
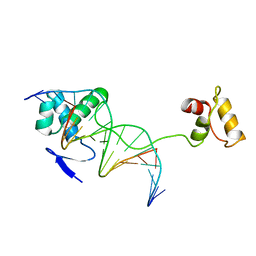 | | CRYSTAL STRUCTURE OF A PAIRED DOMAIN-DNA COMPLEX AT 2.5 ANGSTROMS RESOLUTION REVEALS STRUCTURAL BASIS FOR PAX DEVELOPMENTAL MUTATIONS | | Descriptor: | DNA (5'-D(*AP*AP*CP*GP*TP*CP*AP*CP*GP*GP*TP*TP*GP*AP*C)-3'), DNA (5'-D(*TP*TP*GP*TP*CP*AP*AP*CP*CP*GP*TP*GP*AP*CP*G)-3'), PROTEIN (PRD PAIRED) | | Authors: | Xu, W, Rould, M.A, Jun, S, Desplan, C, Pabo, C.O. | | Deposit date: | 1995-05-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a paired domain-DNA complex at 2.5 A resolution reveals structural basis for Pax developmental mutations.
Cell(Cambridge,Mass.), 80, 1995
|
|
1PFB
 
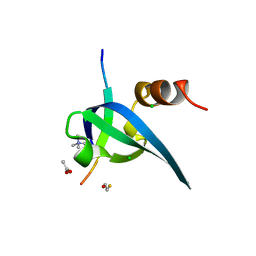 | | Structural Basis for specific binding of polycomb chromodomain to histone H3 methylated at K27 | | Descriptor: | ACETIC ACID, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Min, J.R, Zhang, Y, Xu, R.-M. | | Deposit date: | 2003-05-24 | | Release date: | 2003-10-07 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27.
Genes Dev., 17, 2003
|
|
1PK1
 
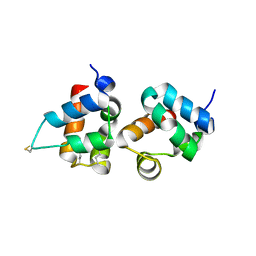 | | Hetero SAM domain structure of Ph and Scm. | | Descriptor: | Polyhomeotic-proximal chromatin protein, Sex comb on midleg CG9495-PA | | Authors: | Kim, C.A, Sawaya, M.R, Cascio, D, Kim, W, Bowie, J.U. | | Deposit date: | 2003-06-04 | | Release date: | 2005-02-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural organization of a Sex-comb-on-midleg/polyhomeotic copolymer.
J.Biol.Chem., 280, 2005
|
|
1PK3
 
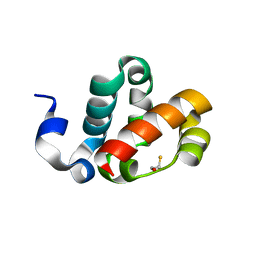 | | Scm SAM domain | | Descriptor: | BETA-MERCAPTOETHANOL, Sex comb on midleg CG9495-PA | | Authors: | Kim, C.A, Sawaya, M.R, Cascio, D, Kim, W, Bowie, J.U. | | Deposit date: | 2003-06-04 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural organization of a Sex-comb-on-midleg/polyhomeotic copolymer.
J.Biol.Chem., 280, 2005
|
|
2DB3
 
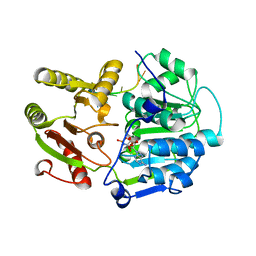 | | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-dependent RNA helicase vasa, MAGNESIUM ION, ... | | Authors: | Sengoku, T, Nureki, O, Nakamura, A, Kobayashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa.
Cell(Cambridge,Mass.), 125, 2006
|
|
2DH1
 
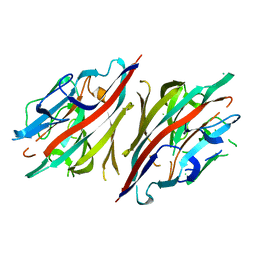 | | Crystal structure of peanut lectin lactose-azobenzene-4,4'-dicarboxylic acid-lactose complex | | Descriptor: | Galactose-binding lectin | | Authors: | Natchiar, S.K, Srinivas, O, Nivedita, M, Sagarika, D, Jayaraman, N, Surolia, A, Vijayan, M. | | Deposit date: | 2006-03-17 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (7.65 Å) | | Cite: | Multivalency in lectins - A crystallographic, modelling and light-scattering study involving peanut lectin and a bivalent ligand
Curr.Sci., 90, 2006
|
|
1NLQ
 
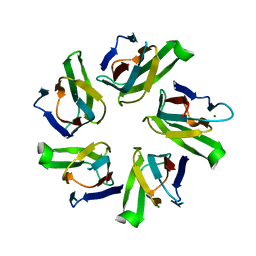 | | The crystal structure of Drosophila NLP-core provides insight into pentamer formation and histone binding | | Descriptor: | MAGNESIUM ION, Nucleoplasmin-like protein | | Authors: | Namboodiri, V.M.H, Dutta, S, Akey, I.V, Head, J.F, Akey, C.W. | | Deposit date: | 2003-01-07 | | Release date: | 2003-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of Drosophila NLP-core Provides Insight into Pentamer Formation and Histone Binding
Structure, 11, 2003
|
|
1ZQ3
 
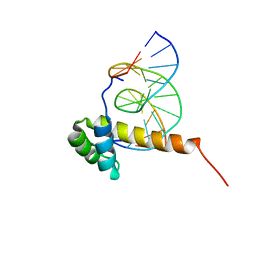 | | NMR Solution Structure of the Bicoid Homeodomain Bound to the Consensus DNA Binding Site TAATCC | | Descriptor: | 5'-D(*CP*GP*GP*GP*GP*AP*TP*TP*AP*GP*AP*GP*C)-3', 5'-D(*GP*CP*TP*CP*TP*AP*AP*TP*CP*CP*CP*CP*G)-3', Homeotic bicoid protein | | Authors: | Baird-Titus, J.M, Rance, M, Clark-Baldwin, K, Ma, J, Vrushank, D. | | Deposit date: | 2005-05-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the native K50 Bicoid homeodomain bound to the consensus TAATCC DNA-binding site.
J.Mol.Biol., 356, 2006
|
|
2DVB
 
 | | Crystal structure of peanut lectin GAl-beta-1,6-GalNAc complex | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Natchiar, S.K, Srinivas, O, Mitra, N, Surolia, A, Jayaraman, N, Vijayan, M. | | Deposit date: | 2006-07-30 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural studies on peanut lectin complexed with disaccharides involving different linkages: further insights into the structure and interactions of the lectin
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2DVF
 
 | | Crystals of peanut lectin grown in the presence of GAL-ALPHA-1,3-GAL-BETA-1,4-GAL | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Natchiar, S.K, Srinivas, O, Mitra, N, Surolia, A, Jayaraman, N, Vijayan, M. | | Deposit date: | 2006-07-31 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural studies on peanut lectin complexed with disaccharides involving different linkages: further insights into the structure and interactions of the lectin
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
