9G6I
 
 | |
1LWF
 
 | | CRYSTAL STRUCTURE OF A MUTANT HIV-1 REVERSE TRANSCRIPTASE (RTMQ+M184V: M41L/D67N/K70R/M184V/T215Y) IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
1LXA
 
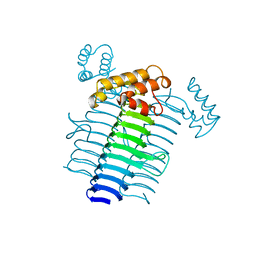 | | UDP N-ACETYLGLUCOSAMINE ACYLTRANSFERASE | | Descriptor: | UDP N-ACETYLGLUCOSAMINE O-ACYLTRANSFERASE | | Authors: | Roderick, S.L. | | Deposit date: | 1995-10-07 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A left-handed parallel beta helix in the structure of UDP-N-acetylglucosamine acyltransferase.
Science, 270, 1995
|
|
9G81
 
 | | CTX-M-14 mixed with piperacillin at 3s delay time - serial crystallography temperature series; 30C, 303K | | Descriptor: | Beta-lactamase, Hydrolyzed piperacillin, Piperacillin, ... | | Authors: | Prester, A, von Stetten, D, Mehrabi, P, Schulz, E.C. | | Deposit date: | 2024-07-22 | | Release date: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Probing the modulation of enzyme kinetics by multi-temperature, time-resolved serial crystallography.
Nat Commun, 16, 2025
|
|
1LY3
 
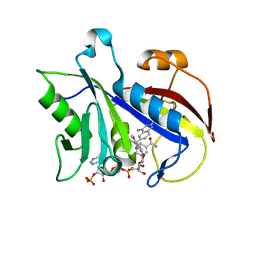 | | ANALYSIS OF QUINAZOLINE AND PYRIDOPYRIMIDINE N9-C10 REVERSED BRIDGE ANTIFOLATES IN COMPLEX WITH NADP+ AND PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE | | Descriptor: | 2,4-DIAMINO-6-[N-(2',5'-DIMETHOXYBENZYL)-N-METHYLAMINO]QUINAZOLINE, DIHYDROFOLATE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Queener, S.F, Gangjee, A. | | Deposit date: | 2002-06-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of quinazoline and pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
9G7X
 
 | | CTX-M-14 apo serial crystallography temperature series; 30C, 303K | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Prester, A, von Stetten, D, Mehrabi, P, Schulz, E.C. | | Deposit date: | 2024-07-22 | | Release date: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the modulation of enzyme kinetics by multi-temperature, time-resolved serial crystallography.
Nat Commun, 16, 2025
|
|
1LYJ
 
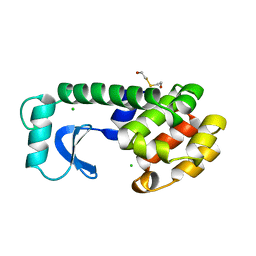 | | DISSECTION OF HELIX CAPPING IN T4 LYSOZYME BY STRUCTURAL AND THERMODYNAMIC ANALYSIS OF SIX AMINO ACID SUBSTITUTIONS AT THR 59 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Becktel, W.J, Sauer, U, Baase, W.A, Matthews, B.W. | | Deposit date: | 1992-08-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissection of helix capping in T4 lysozyme by structural and thermodynamic analysis of six amino acid substitutions at Thr 59.
Biochemistry, 31, 1992
|
|
9G1K
 
 | | Fragment screening of FosAKP, cryo structure in complex with fragment F2X-entry A09 | | Descriptor: | 1,2-ETHANEDIOL, 2-sulfanylpyridine-3-carboximidamide, Fosfomycin resistance protein, ... | | Authors: | Guenther, S, Galchenkova, M, Fischer, P, Reinke, P.Y.A, Falke, S, Thekku Veedu, S, Rodrigues, A.C, Senst, J, Meents, A. | | Deposit date: | 2024-07-10 | | Release date: | 2025-07-23 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Room-temperature X-ray fragment screening with serial crystallography.
Nat Commun, 16, 2025
|
|
1MQI
 
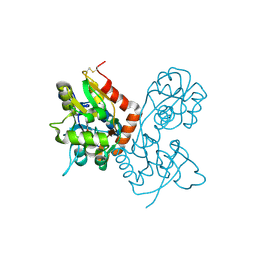 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Fluoro-Willardiine at 1.35 Angstroms Resolution | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
9FY7
 
 | | Dye Type Peroxidase Aa from Streptomyces lividans with N3 ligand by serial electron diffraction (SerialED) | | Descriptor: | AZIDE ION, Deferrochelatase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hofer, G, Wang, L, Pacoste, L, Hager, P, Finjallaz, A, Williams, L, Worral, J, Steiner, R, Xu, H, Zou, X. | | Deposit date: | 2024-07-03 | | Release date: | 2025-07-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | Cite: | Universal Serial electron diffraction for high quality protein structures
To Be Published
|
|
9FYK
 
 | | Dye Type Peroxidase Aa from Streptomyces lividans by serial electron diffraction (SerialED) | | Descriptor: | Deferrochelatase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hofer, G, Wang, L, Pacoste, L, Hager, P, Finjallaz, A, Williams, L, Worral, J, Steiner, R, Xu, H, Zou, X. | | Deposit date: | 2024-07-03 | | Release date: | 2025-07-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.3 Å) | | Cite: | Universal Serial electron diffraction for high quality protein structures
To Be Published
|
|
9FXM
 
 | | TRPC4 in complex with Z-AzPico | | Descriptor: | (Z)-7-(4-chlorobenzyl)-1-(3-hydroxypropyl)-3-methyl-8-(4-(phenyldiazenyl)-3-(trifluoromethoxy)phenoxy)-3,7-dihydro-1H-purine-2,6-dione, Transient receptor potential cation channel subfamily c member 4a | | Authors: | Vinayagam, D, Raunser, S. | | Deposit date: | 2024-07-01 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | TRPC4 in complex with Z-AzPico
To Be Published
|
|
1LYS
 
 | |
1LZ4
 
 | | ENTHALPIC DESTABILIZATION OF A MUTANT HUMAN LYSOZYME LACKING A DISULFIDE BRIDGE BETWEEN CYSTEINE-77 AND CYSTEINE-95 | | Descriptor: | HUMAN LYSOZYME | | Authors: | Inaka, K, Matsushima, M, Kuroki, R, Taniyama, Y, Kikuchi, M. | | Deposit date: | 1993-02-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enthalpic destabilization of a mutant human lysozyme lacking a disulfide bridge between cysteine-77 and cysteine-95.
Biochemistry, 31, 1992
|
|
9GJ3
 
 | | Structure of the amyloid-forming peptide LYIQNY | | Descriptor: | Peptide LYIQNY | | Authors: | Durvanger, Z. | | Deposit date: | 2024-08-21 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Chemical Evolution of Early Macromolecules: From Prebiotic Oligopeptides to Self-Organizing Biosystems via Amyloid Formation.
Chemistry, 31, 2025
|
|
1LZ9
 
 | |
9G1C
 
 | | Fragment screening of FosAKP, room-temperature structure in complex with fragment F2X-entry A12 | | Descriptor: | 3-(propan-2-yl)-1,2,4-oxadiazol-5(4H)-one, Fosfomycin resistance protein, MANGANESE (II) ION | | Authors: | Guenther, S, Galchenkova, M, Fischer, P, Reinke, P.Y.A, Falke, S, Thekku Veedu, S, Rodrigues, A.C, Senst, J, Meents, A. | | Deposit date: | 2024-07-10 | | Release date: | 2025-07-23 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Room-temperature X-ray fragment screening with serial crystallography.
Nat Commun, 16, 2025
|
|
9G1J
 
 | | Fragment screening of FosAKP, cryo structure in complex with fragment F2X-entry A06 | | Descriptor: | 1,2-ETHANEDIOL, 3-methylsulfanyl-1,2,4-triazine, Fosfomycin resistance protein, ... | | Authors: | Guenther, S, Galchenkova, M, Fischer, P, Reinke, P.Y.A, Falke, S, Thekku Veedu, S, Rodrigues, A.C, Senst, J, Meents, A. | | Deposit date: | 2024-07-10 | | Release date: | 2025-07-23 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Room-temperature X-ray fragment screening with serial crystallography.
Nat Commun, 16, 2025
|
|
9G1A
 
 | | Fragment screening of FosAKP, room-temperature structure, ground state | | Descriptor: | Fosfomycin resistance protein, MANGANESE (II) ION | | Authors: | Guenther, S, Galchenkova, M, Fischer, P, Reinke, P.Y.A, Falke, S, Thekku Veedu, S, Rodrigues, A.C, Senst, J, Meents, A. | | Deposit date: | 2024-07-10 | | Release date: | 2025-07-23 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Room-temperature X-ray fragment screening with serial crystallography.
Nat Commun, 16, 2025
|
|
1LWA
 
 | | Solution Structure of SRY_DNA | | Descriptor: | 5'-D(*CP*TP*GP*AP*AP*CP*AP*AP*TP*CP*AP*CP*CP*CP*C)-3', 5'-D(*GP*GP*GP*GP*TP*GP*AP*TP*TP*GP*TP*TP*CP*AP*G)-3' | | Authors: | Masse, J.E, Wong, B, Yen, Y.-M, Allain, F.H.-T, Johnson, R.C, Feigon, J. | | Deposit date: | 2002-05-30 | | Release date: | 2002-10-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding
J.Mol.Biol., 323, 2002
|
|
1LWT
 
 | |
9G1G
 
 | | Fragment screening of FosAKP, room-temperature structure in complex with fragment F2X-entry G08 | | Descriptor: | Fosfomycin resistance protein, MANGANESE (II) ION, N-ethyl-2-{[5-(propan-2-yl)-1,3,4-oxadiazol-2-yl]sulfanyl}acetamide | | Authors: | Guenther, S, Galchenkova, M, Fischer, P, Reinke, P.Y.A, Falke, S, Thekku Veedu, S, Rodrigues, A.C, Senst, J, Meents, A. | | Deposit date: | 2024-07-10 | | Release date: | 2025-07-23 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Room-temperature X-ray fragment screening with serial crystallography.
Nat Commun, 16, 2025
|
|
1LXH
 
 | |
9G1N
 
 | | Fragment screening of FosAKP, cryo structure in complex with fragment F2X-entry E04 | | Descriptor: | 1,2-ETHANEDIOL, 3-phenyl-1,2-oxazol-5-amine, Fosfomycin resistance protein, ... | | Authors: | Guenther, S, Galchenkova, M, Fischer, P, Reinke, P.Y.A, Falke, S, Thekku Veedu, S, Rodrigues, A.C, Senst, J, Meents, A. | | Deposit date: | 2024-07-10 | | Release date: | 2025-07-23 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Room-temperature X-ray fragment screening with serial crystallography.
Nat Commun, 16, 2025
|
|
1LY1
 
 | |
