1FU8
 
 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | 1-DEOXY-1-ACETYLAMINO-BETA-D-GLUCO-2-HEPTULOPYRANOSONAMIDE, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-14 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
1FU9
 
 | | SOLUTION STRUCTURE OF THE NINTH ZINC-FINGER DOMAIN OF THE U-SHAPED TRANSCRIPTION FACTOR | | Descriptor: | U-SHAPED TRANSCRIPTIONAL COFACTOR, ZINC ION | | Authors: | Liew, C.K, Kowalski, K, Fox, A.H, Newton, A, Sharpe, B.K, Crossley, M, Mackay, J.P. | | Deposit date: | 2000-09-14 | | Release date: | 2000-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two CCHC zinc fingers from the FOG family protein U-shaped that mediate protein-protein interactions.
Structure Fold.Des., 8, 2000
|
|
1FUA
 
 | | L-FUCULOSE 1-PHOSPHATE ALDOLASE CRYSTAL FORM T | | Descriptor: | BETA-MERCAPTOETHANOL, L-FUCULOSE-1-PHOSPHATE ALDOLASE, SULFATE ION, ... | | Authors: | Dreyer, M.K, Schulz, G.E. | | Deposit date: | 1996-02-14 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Refined high-resolution structure of the metal-ion dependent L-fuculose-1-phosphate aldolase (class II) from Escherichia coli.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1FUB
 
 | | FIRST PROTEIN STRUCTURE DETERMINED FROM X-RAY POWDER DIFFRACTION DATA | | Descriptor: | CHLORIDE ION, INSULIN, A CHAIN, ... | | Authors: | Von Dreele, R.B, Stephens, P.W, Blessing, R.H, Smith, G.D. | | Deposit date: | 2000-09-14 | | Release date: | 2000-10-16 | | Last modified: | 2024-10-30 | | Method: | POWDER DIFFRACTION | | Cite: | The first protein crystal structure determined from high-resolution X-ray powder diffraction data: a variant of T3R3 human insulin-zinc complex produced by grinding.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1FUE
 
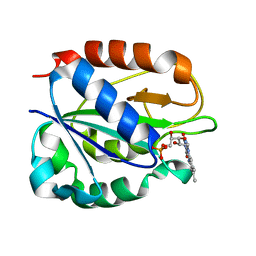 | | FLAVODOXIN FROM HELICOBACTER PYLORI | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Freigang, J, Diederichs, K, Schaefer, K.P, Welte, W, Paul, R. | | Deposit date: | 2000-09-15 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of oxidized flavodoxin, an essential protein in Helicobacter pylori.
Protein Sci., 11, 2002
|
|
1FUF
 
 | | CRYSTAL STRUCTURE OF A 14BP RNA OLIGONUCLEOTIDE CONTAINING DOUBLE UU BULGES: A NOVEL INTRAMOLECULAR U*(AU) BASE TRIPLE | | Descriptor: | (5'-R(*GP*GP*UP*AP*UP*UP*UP*CP*GP*GP*UP*AP*(CBR)P*C)-3'), MAGNESIUM ION, SPERMINE | | Authors: | Deng, J, Xiong, Y, Sudarsanakumar, C, Shi, K, Sundaralingam, M. | | Deposit date: | 2000-09-15 | | Release date: | 2001-11-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of two forms of a 14-mer RNA/DNA chimer duplex with double UU bulges: a novel intramolecular U*(A x U) base triple.
RNA, 7, 2001
|
|
1FUG
 
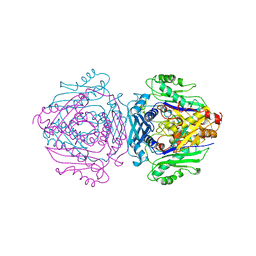 | | S-ADENOSYLMETHIONINE SYNTHETASE | | Descriptor: | S-ADENOSYLMETHIONINE SYNTHETASE | | Authors: | Fu, Z, Markham, G.D, Takusagawa, F. | | Deposit date: | 1996-02-25 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Flexible loop in the structure of S-adenosylmethionine synthetase crystallized in the tetragonal modification.
J.Biomol.Struct.Dyn., 13, 1996
|
|
1FUI
 
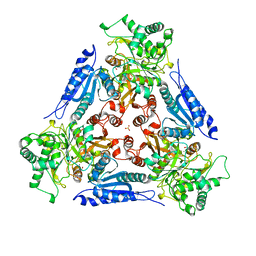 | | L-FUCOSE ISOMERASE FROM ESCHERICHIA COLI | | Descriptor: | FUCITOL, L-FUCOSE ISOMERASE, MANGANESE (II) ION, ... | | Authors: | Seemann, J.E, Schulz, G.E. | | Deposit date: | 1997-04-14 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of L-fucose isomerase from Escherichia coli.
J.Mol.Biol., 273, 1997
|
|
1FUJ
 
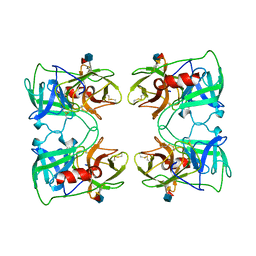 | | PR3 (MYELOBLASTIN) | | Descriptor: | PR3, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Fujinaga, M, Chernaia, M.M, Halenbeck, R, Koths, K, James, M.N.G. | | Deposit date: | 1996-01-25 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of PR3, a neutrophil serine proteinase antigen of Wegener's granulomatosis antibodies.
J.Mol.Biol., 261, 1996
|
|
1FUK
 
 | |
1FUL
 
 | |
1FUN
 
 | | SUPEROXIDE DISMUTASE MUTANT WITH LYS 136 REPLACED BY GLU, CYS 6 REPLACED BY ALA AND CYS 111 REPLACED BY SER (K136E, C6A, C111S) | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE, ... | | Authors: | Lo, T.P, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Computational, pulse-radiolytic, and structural investigations of lysine-136 and its role in the electrostatic triad of human Cu,Zn superoxide dismutase.
Proteins, 29, 1997
|
|
1FUO
 
 | | FUMARASE C WITH BOUND CITRATE | | Descriptor: | CITRIC ACID, D-MALATE, FUMARASE C | | Authors: | Weaver, T, Banaszak, L. | | Deposit date: | 1996-08-29 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystallographic studies of the catalytic and a second site in fumarase C from Escherichia coli.
Biochemistry, 35, 1996
|
|
1FUP
 
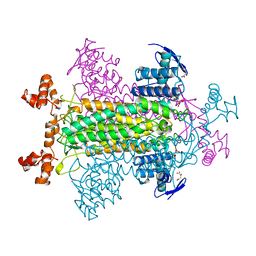 | | FUMARASE WITH BOUND PYROMELLITIC ACID | | Descriptor: | D-MALATE, FUMARASE C, PYROMELLITIC ACID | | Authors: | Weaver, T, Banaszak, L. | | Deposit date: | 1996-08-29 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic studies of the catalytic and a second site in fumarase C from Escherichia coli.
Biochemistry, 35, 1996
|
|
1FUQ
 
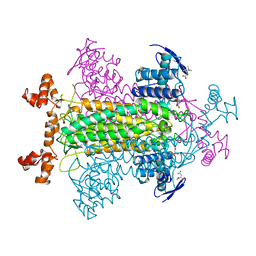 | | FUMARASE WITH BOUND 3-TRIMETHYLSILYLSUCCINIC ACID | | Descriptor: | 3-TRIMETHYLSILYLSUCCINIC ACID, CITRIC ACID, FUMARASE C | | Authors: | Weaver, T, Banaszak, L. | | Deposit date: | 1996-08-29 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies of the catalytic and a second site in fumarase C from Escherichia coli.
Biochemistry, 35, 1996
|
|
1FUR
 
 | |
1FUS
 
 | |
1FUT
 
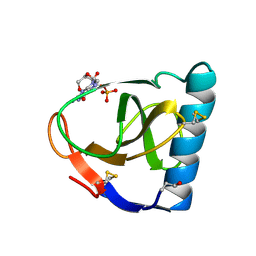 | | CRYSTAL STRUCTURES OF RIBONUCLEASE F1 OF FUSARIUM MONILIFORME IN ITS FREE FORM AND IN COMPLEX WITH 2'GMP | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE F1 | | Authors: | Katayanagi, K, Vassylyev, D.G, Ishikawa, K, Morikawa, K. | | Deposit date: | 1993-01-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of ribonuclease F1 of Fusarium moniliforme in its free form and in complex with 2'GMP.
J.Mol.Biol., 230, 1993
|
|
1FUU
 
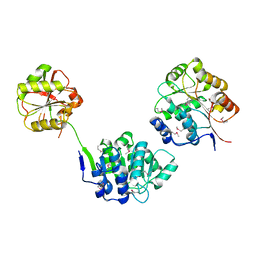 | | YEAST INITIATION FACTOR 4A | | Descriptor: | YEAST INITIATION FACTOR 4A | | Authors: | Caruthers, J.M, Johnson, E.R, McKay, D.B. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FUV
 
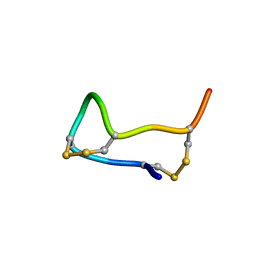 | |
1FUW
 
 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF A DOUBLE MUTANT SINGLE-CHAIN MONELLIN(SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | MONELLIN | | Authors: | Sung, Y.H, Shin, J, Jung, J, Lee, W. | | Deposit date: | 2000-09-18 | | Release date: | 2001-06-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure, backbone dynamics, and stability of a double mutant single-chain monellin. structural origin of sweetness.
J.Biol.Chem., 276, 2001
|
|
1FUX
 
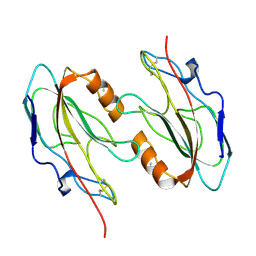 | | CRYSTAL STRUCTURE OF E.COLI YBCL, A NEW MEMBER OF THE MAMMALIAN PEBP FAMILY | | Descriptor: | HYPOTHETICAL 19.5 KDA PROTEIN IN EMRE-RUS INTERGENIC REGION | | Authors: | Serre, L, Pereira de Jesus, K, Benedetti, H, Bureaud, N, Schoentgen, F, Zelwer, C. | | Deposit date: | 2000-09-18 | | Release date: | 2001-07-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of YBHB and YBCL from Escherichia coli, two bacterial homologues to a Raf kinase inhibitor protein.
J.Mol.Biol., 310, 2001
|
|
1FUY
 
 | |
1FV0
 
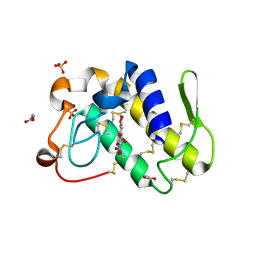 | | FIRST STRUCTURAL EVIDENCE OF THE INHIBITION OF PHOSPHOLIPASE A2 BY ARISTOLOCHIC ACID: CRYSTAL STRUCTURE OF A COMPLEX FORMED BETWEEN PHOSPHOLIPASE A2 AND ARISTOLOCHIC ACID | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 9-HYDROXY ARISTOLOCHIC ACID, ACETATE ION, ... | | Authors: | Chandra, V, Jasti, J, Kaur, P, Srinivasan, A, Betzel, C, Singh, T.P. | | Deposit date: | 2000-09-18 | | Release date: | 2002-08-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Phospholipase A2 Inhibition for the Synthesis of Prostaglandins by the Plant Alkaloid Aristolochic Acid from a 1.7 A Crystal Structure
Biochemistry, 41, 2002
|
|
1FV1
 
 | | STRUCTURAL BASIS FOR THE BINDING OF AN IMMUNODOMINANT PEPTIDE FROM MYELIN BASIC PROTEIN IN DIFFERENT REGISTERS BY TWO HLA-DR2 ALLELES | | Descriptor: | GLYCEROL, MAJOR HISTOCOMPATIBILITY COMPLEX ALPHA CHAIN, MAJOR HISTOCOMPATIBILITY COMPLEX BETA CHAIN, ... | | Authors: | Li, H, Mariuzza, A.R, Li, Y, Martin, R. | | Deposit date: | 2000-09-18 | | Release date: | 2000-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the binding of an immunodominant peptide from myelin basic protein in different registers by two HLA-DR2 proteins.
J.Mol.Biol., 304, 2000
|
|
