9G5H
 
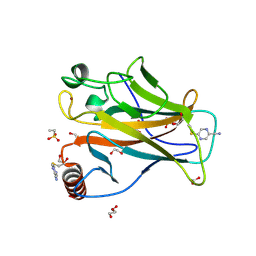 | | p53-Y220C Core Domain Covalently Bound to 2-chloro-5-cyanopyrazine Soaked at 5 mM | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-chloranylpyrazine-2-carbonitrile, ... | | Authors: | Stahlecker, J, Klett, T, Stehle, T, Boeckler, F.M. | | Deposit date: | 2024-07-17 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | S N Ar Reactive Pyrazine Derivatives as p53-Y220C Cleft Binders with Diverse Binding Modes.
Drug Des Devel Ther, 19, 2025
|
|
1MQR
 
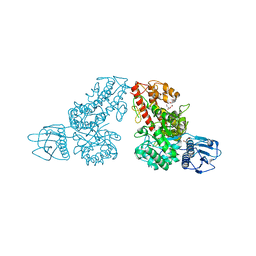 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE (E386Q) FROM BACILLUS STEAROTHERMOPHILUS T-6 | | Descriptor: | ALPHA-D-GLUCURONIDASE, GLYCEROL | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
9G6U
 
 | | p53-Y220C Core Domain Covalently Bound to 3,5-Dichloro-6-Ethylpyrazine-2-carbonitirle Soaked at 5 mM | | Descriptor: | 1,2-ETHANEDIOL, 3,5-bis(chloranyl)-6-ethyl-pyrazine-2-carbonitrile, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Stahlecker, J, Klett, T, Stehle, T, Boeckler, F.M. | | Deposit date: | 2024-07-19 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | S N Ar Reactive Pyrazine Derivatives as p53-Y220C Cleft Binders with Diverse Binding Modes.
Drug Des Devel Ther, 19, 2025
|
|
1LV0
 
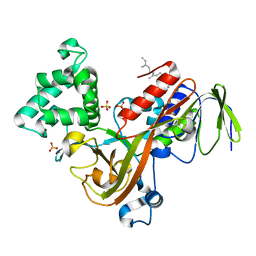 | | Crystal structure of the Rab effector guanine nucleotide dissociation inhibitor (GDI) in complex with a geranylgeranyl (GG) peptide | | Descriptor: | GERAN-8-YL GERAN, RAB GDP disossociation inhibitor alpha, SULFATE ION | | Authors: | An, Y, Shao, Y, Alory, C, Matteson, J, Sakisaka, T, Chen, W, Gibbs, R.A, Wilson, I.A, Balch, W.E. | | Deposit date: | 2002-05-23 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Geranylgeranyl switching regulates GDI-Rab GTPase recycling.
Structure, 11, 2003
|
|
9GD8
 
 | | NME1 94-Diphosphoserine | | Descriptor: | Nucleoside diphosphate kinase A | | Authors: | Roderer, D, Schoepf, F, Fiedler, D, Celik, A. | | Deposit date: | 2024-08-05 | | Release date: | 2025-06-11 | | Last modified: | 2025-09-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Nucleoside diphosphate kinase A (NME1) catalyses its own oligophosphorylation.
Nat.Chem., 2025
|
|
9G27
 
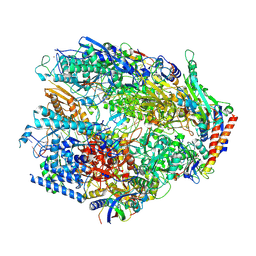 | | Yeast RNA polymerase I elongation complex stalled by an apurinic site, pre-translocation state | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Santos-Aledo, A, Plaza-Pegueroles, A, Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM uncovers a sequential mechanism for RNA polymerase I pausing and stalling at abasic DNA lesions.
Nat Commun, 16, 2025
|
|
7RKB
 
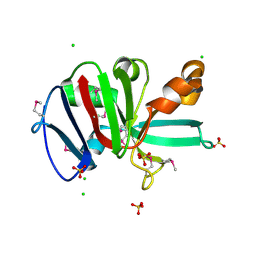 | | Crystal Structure of Putative Pterin Binding Protein (PruR) from Klebsiella pneumoniae in Complex with Neopterin | | Descriptor: | CHLORIDE ION, L-NEOPTERIN, Pterin Binding Protein, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Putative Pterin Binding Protein (PruR) from Klebsiella pneumoniae in Complex with Neopterin.
To Be Published
|
|
1MM9
 
 | | Streptavidin Mutant with Insertion of Fibronectin Hexapeptide, including RGD | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Streptavidin | | Authors: | Le Trong, I, McDevitt, T.C, Nelson, K.E, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-09-03 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural characterization and comparison of RGD cell-adhesion recognition sites engineered into streptavidin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1MMC
 
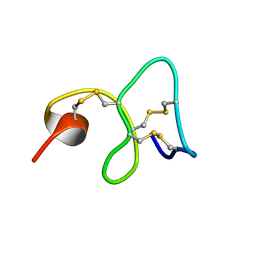 | | 1H NMR STUDY OF THE SOLUTION STRUCTURE OF AC-AMP2 | | Descriptor: | ANTIMICROBIAL PEPTIDE 2 | | Authors: | Martins, J.C, Maes, D, Loris, R, Pepermans, H.A.M, Wyns, L, Willem, R, Verheyden, P. | | Deposit date: | 1995-10-25 | | Release date: | 1996-03-08 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | H NMR study of the solution structure of Ac-AMP2, a sugar binding antimicrobial protein isolated from Amaranthus caudatus.
J.Mol.Biol., 258, 1996
|
|
9G1X
 
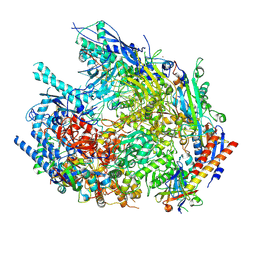 | | Yeast RNA polymerase I elongation complex stalled by an apurinic site, 11-subunit | | Descriptor: | DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, DNA-directed RNA polymerase I subunit RPA190, ... | | Authors: | Santos-Aledo, A, Plaza-Pegueroles, A, Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM uncovers a sequential mechanism for RNA polymerase I pausing and stalling at abasic DNA lesions.
Nat Commun, 16, 2025
|
|
9G2B
 
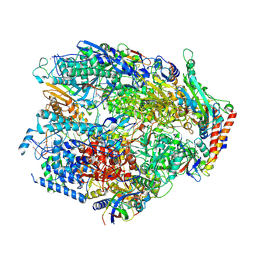 | | Yeast RNA polymerase I elongation complex stalled by an apurinic site, 12-subunit | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Santos-Aledo, A, Plaza-Pegueroles, A, Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM uncovers a sequential mechanism for RNA polymerase I pausing and stalling at abasic DNA lesions.
Nat Commun, 16, 2025
|
|
2PEH
 
 | | Crystal structure of the UHM domain of human SPF45 in complex with SF3b155-ULM5 | | Descriptor: | Splicing factor 3B subunit 1, Splicing factor 45 | | Authors: | Corsini, L, Basquin, J, Hothorn, M, Sattler, M. | | Deposit date: | 2007-04-03 | | Release date: | 2007-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | U2AF-homology motif interactions are required for alternative splicing regulation by SPF45.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1MMW
 
 | | Rat neuronal NOS heme domain with vinyl-L-NIO bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N5-(1-IMINO-3-BUTENYL)-L-ORNITHINE, ... | | Authors: | Bretscher, L.E, Li, H, Poulos, T.L, Griffith, O.W. | | Deposit date: | 2002-09-04 | | Release date: | 2003-09-16 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization and kinetics of nitric-oxide synthase inhibition by novel N5-(iminoalkyl)- and N5-(iminoalkenyl)-ornithines
J.Biol.Chem., 278, 2003
|
|
9G23
 
 | | Yeast RNA polymerase I elongation complex stalled by an apurinic site bound to nucleotide analog AMPCPP at A-site | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, ... | | Authors: | Santos-Aledo, A, Plaza-Pegueroles, A, Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM uncovers a sequential mechanism for RNA polymerase I pausing and stalling at abasic DNA lesions.
Nat Commun, 16, 2025
|
|
1MNA
 
 | | Thioesterase Domain of Picromycin Polyketide Synthase (PICS TE), pH 8.0 | | Descriptor: | polyketide synthase IV | | Authors: | Tsai, S.-C, Lu, H, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2002-09-05 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into channel architecture and substrate specificity from crystal structures of two macrocycle-forming thioesterases of modular polyketide synthases
Biochemistry, 41, 2002
|
|
7ROM
 
 | |
9G29
 
 | | Yeast RNA polymerase I elongation complex stalled by an apurinic site with the C-terminal of A12 in the funnel | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Santos-Aledo, A, Plaza-Pegueroles, A, Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM uncovers a sequential mechanism for RNA polymerase I pausing and stalling at abasic DNA lesions.
Nat Commun, 16, 2025
|
|
9G24
 
 | | Yeast RNA polymerase I elongation complex stalled by an apurinic site bound to nucleotide analog AMPCPP at E-site | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, ... | | Authors: | Santos-Aledo, A, Plaza-Pegueroles, A, Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM uncovers a sequential mechanism for RNA polymerase I pausing and stalling at abasic DNA lesions.
Nat Commun, 16, 2025
|
|
1LX6
 
 | |
9GBY
 
 | | Crystal structure of Pseudomonas aeruginosa IspD | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, POTASSIUM ION | | Authors: | Borel, F, D'Auria, L. | | Deposit date: | 2024-07-31 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Fragment Discovery by X-Ray Crystallographic Screening Targeting the CTP Binding Site of Pseudomonas Aeruginosa IspD.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
7RHP
 
 | |
1MNQ
 
 | | Thioesterase Domain of Picromycin Polyketide Synthase (PICS TE), pH 8.4 | | Descriptor: | polyketide synthase IV | | Authors: | Tsai, S.-C, Lu, H, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2002-09-05 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into channel architecture and substrate specificity from crystal structures of two macrocycle-forming thioesterases of modular polyketide synthases
Biochemistry, 41, 2002
|
|
1MO8
 
 | | ATPase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Sodium/Potassium-Transporting ATPase alpha-1 | | Authors: | Hilge, M, Siegal, G, Vuister, G.W, Guentert, P, Gloor, S.M, Abrahams, J.P. | | Deposit date: | 2002-09-08 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ATP-induced conformational changes of the nucleotide-binding domain of Na,K-ATPase
Nat.Struct.Biol., 10, 2003
|
|
9GTP
 
 | | Cryo-EM structure of a contractile injection system in Streptomyces coelicolor, the baseplate complex in extended state applied 6-fold symmetry. | | Descriptor: | Baseplate protein J-like domain-containing protein, IraD/Gp25-like domain-containing protein, LysM domain-containing protein, ... | | Authors: | Casu, B, Sallmen, J.W, Haas, P.E, Afanasyev, P, Xu, J, Schlimpert, S, Pilhofer, M. | | Deposit date: | 2024-09-18 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Function and firing of the Streptomyces coelicolor contractile injection system requires the membrane protein CisA.
Elife, 14, 2025
|
|
9G2C
 
 | | Yeast RNA polymerase I elongation complex stalled by an apurinic site, open state | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA190, ... | | Authors: | Santos-Aledo, A, Plaza-Pegueroles, A, Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM uncovers a sequential mechanism for RNA polymerase I pausing and stalling at abasic DNA lesions.
Nat Commun, 16, 2025
|
|
