4J6K
 
 | |
4J6Q
 
 | |
4J6J
 
 | |
4J6N
 
 | |
4J6M
 
 | |
4J6P
 
 | |
4J6L
 
 | |
4JPI
 
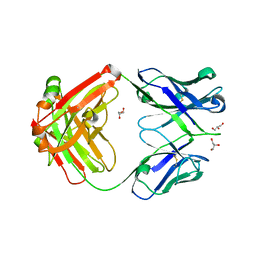 | | Crystal structure of a putative VRC01 germline precursor Fab | | Descriptor: | GLYCEROL, Putative VRC01 germline Fab heavy chain, Putative VRC01 germline Fab light chain | | Authors: | Julien, J.-P, Diwanji, D.C, Jardine, J, Schief, W.R, Wilson, I.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational HIV immunogen design to target specific germline B cell receptors.
Science, 340, 2013
|
|
8TGO
 
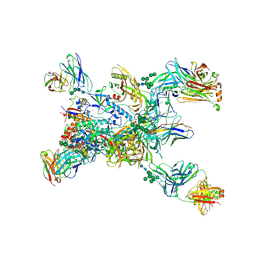 | | Crystal structure of the BG505 triple tandem trimer gp140 HIV-1 Env in complex with PGT124 and 35O22 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 scFv, ... | | Authors: | Xian, Y, Yuan, M, Wilson, I.A. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (5.75 Å) | | Cite: | Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines.
Npj Vaccines, 9, 2024
|
|
4NCO
 
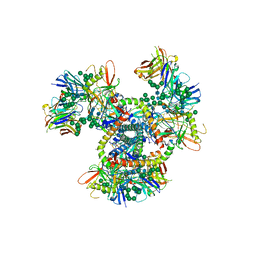 | | Crystal Structure of the BG505 SOSIP gp140 HIV-1 Env trimer in Complex with the Broadly Neutralizing Fab PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 SOSIP gp120, BG505 SOSIP gp41, ... | | Authors: | Julien, J.-P, Stanfield, R.L, Lyumkis, D, Ward, A.B, Wilson, I.A. | | Deposit date: | 2013-10-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.7 Å) | | Cite: | Crystal structure of a soluble cleaved HIV-1 envelope trimer.
Science, 342, 2013
|
|
5UM8
 
 | | Crystal structure of HIV-1 envelope trimer 16055 NFL TD CC (T569G) in complex with Fabs 35022 and PGT124 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 35022 heavy chain, ... | | Authors: | Garces, F, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.935 Å) | | Cite: | Glycine Substitution at Helix-to-Coil Transitions Facilitates the Structural Determination of a Stabilized Subtype C HIV Envelope Glycoprotein.
Immunity, 46, 2017
|
|
5VF6
 
 | |
7SOD
 
 | |
7SOB
 
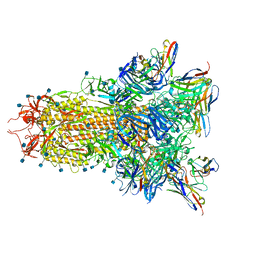 | |
7SO9
 
 | |
7SOC
 
 | |
7SOE
 
 | |
7SOF
 
 | |
7SOA
 
 | |
6D2P
 
 | |
8CWU
 
 | |
6VYV
 
 | |
6W09
 
 | |
6W1C
 
 | |
8TXT
 
 | | Crystal structure of 05.GC.w13.02 Fab in complex with H5 HA from A/Viet Nam/1203/2004(H5N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GC_W13B_B, ... | | Authors: | Lin, T.H, Moore, N, Wilson, I.A. | | Deposit date: | 2023-08-24 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Maturation of germinal center B cells after influenza virus vaccination in humans.
J.Exp.Med., 221, 2024
|
|
