1M99
 
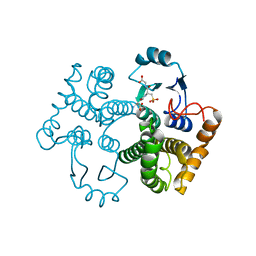 | | Crystal structure of the 26 kDa glutathione S-transferase from Schistosoma japonicum complexed with glutathione sulfonic acid | | Descriptor: | GLUTATHIONE SULFONIC ACID, Glutathione S-Transferase 26kDa | | Authors: | Cardoso, R.M.F, Daniels, D.S, Bruns, C.M, Tainer, J.A. | | Deposit date: | 2002-07-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the electrophile
binding site and substrate binding
mode of the 26-kDa glutathione
S-transferase from Schistosoma
japonicum
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1M9D
 
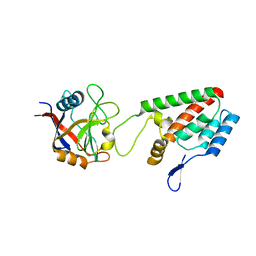 | | X-ray crystal structure of Cyclophilin A/HIV-1 CA N-terminal domain (1-146) O-type chimera Complex. | | Descriptor: | Cyclophilin A, HIV-1 Capsid | | Authors: | Howard, B.R, Vajdos, F.F, Li, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2002-07-28 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the catalytic mechanism of cyclophilin A
Nat.Struct.Biol., 10, 2003
|
|
9GA3
 
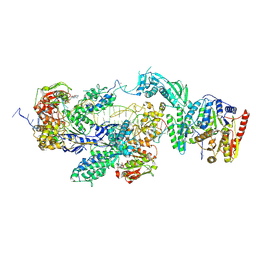 | | MtUvrA2UvrB bound to damaged oligonucleotide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA, UvrABC system protein A, ... | | Authors: | Genta, M, Capelli, R, Ferrara, G, Rizzi, M, Rossi, F, Jeruzalmi, D, Bolognesi, M, Chaves-Sanjuan, A, Miggiano, R. | | Deposit date: | 2024-07-26 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanistic understanding of UvrA damage detection and lesion hand-off to UvrB in Nucleotide Excision Repair.
Nat Commun, 16, 2025
|
|
1LYH
 
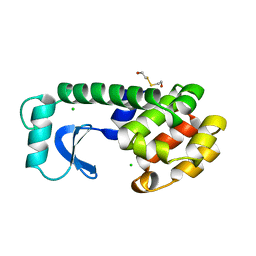 | | DISSECTION OF HELIX CAPPING IN T4 LYSOZYME BY STRUCTURAL AND THERMODYNAMIC ANALYSIS OF SIX AMINO ACID SUBSTITUTIONS AT THR 59 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Becktel, W.J, Sauer, U, Baase, W.A, Matthews, B.W. | | Deposit date: | 1992-08-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dissection of helix capping in T4 lysozyme by structural and thermodynamic analysis of six amino acid substitutions at Thr 59.
Biochemistry, 31, 1992
|
|
9G1Z
 
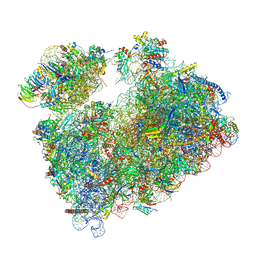 | | Structure of Candida albicans 80S ribosome in complex with mefloquine (non-rotated state) | | Descriptor: | (11R,12S)- Mefloquine, 18S rRNA, 25S rRNA, ... | | Authors: | Kolosova, O, Zgadzay, Y, Stetsenko, A, Atamas, A, Jenner, L.B, Guskov, A, Yusupov, M. | | Deposit date: | 2024-07-10 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of read-through enhancement by aminoglycosides and mefloquine.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9G5E
 
 | | Translation-initiation state of human mitochondrial ribosome small subunit (State F) | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Finke, A.F, Heinrichs, M, Aibara, S, Richter-Dennerlein, R, Hillen, H.S. | | Deposit date: | 2024-07-16 | | Release date: | 2025-04-23 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Coupling of ribosome biogenesis and translation initiation in human mitochondria.
Nat Commun, 16, 2025
|
|
1LYY
 
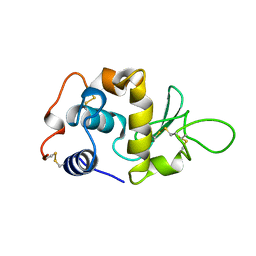 | |
9GLN
 
 | | Crystal Structure of UFC1 C116E | | Descriptor: | GLYCEROL, SULFATE ION, Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Kumar, M, Banerjee, S, Wiener, R. | | Deposit date: | 2024-08-27 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | UFC1 reveals the multifactorial and plastic nature of oxyanion holes in E2 conjugating enzymes.
Nat Commun, 16, 2025
|
|
9GLT
 
 | |
1MA6
 
 | |
9GMM
 
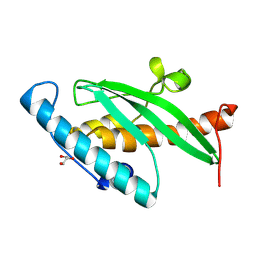 | | Crystal Structure of UFC1 T106I | | Descriptor: | GLYCEROL, Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Kumar, M, Banerjee, S, Wiener, R. | | Deposit date: | 2024-08-29 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | UFC1 reveals the multifactorial and plastic nature of oxyanion holes in E2 conjugating enzymes.
Nat Commun, 16, 2025
|
|
1MB4
 
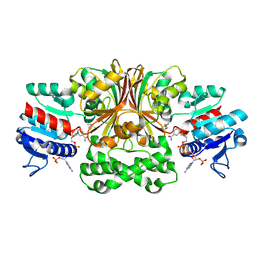 | | Crystal structure of aspartate semialdehyde dehydrogenase from vibrio cholerae with NADP and S-methyl-l-cysteine sulfoxide | | Descriptor: | Aspartate-Semialdehyde Dehydrogenase, CYSTEINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Blanco, J, Moore, R.A, Kabaleeswaran, V, Viola, R.E. | | Deposit date: | 2002-08-02 | | Release date: | 2003-01-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A structural Basis for the Mechanism of Aspartate-beta-semialdehyde Dehydrogenase from Vibrio Cholerae
Protein Sci., 12, 2003
|
|
9GLP
 
 | | Crystal Structure of UFC1 C116E&T106I | | Descriptor: | GLYCEROL, SULFATE ION, Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Kumar, M, Banerjee, S, Wiener, R. | | Deposit date: | 2024-08-27 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | UFC1 reveals the multifactorial and plastic nature of oxyanion holes in E2 conjugating enzymes.
Nat Commun, 16, 2025
|
|
1MBU
 
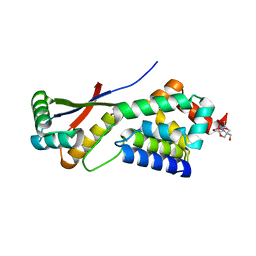 | | Crystal Structure Analysis of ClpSN heterodimer | | Descriptor: | ATP-Dependent clp Protease ATP-Binding Subunit clp A, BIS-(2-HYDROXYETHYL)AMINO-TRIS(HYDROXYMETHYL)METHANE YTTRIUM, CHLORIDE ION, ... | | Authors: | Guo, F, Esser, L, Singh, S.K, Maurizi, M.R, Xia, D. | | Deposit date: | 2002-08-03 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Heterodimeric Complex of the Adaptor, ClpS, with the N-domain of the AAA+ Chaperone, ClpA
J.Biol.Chem., 277, 2002
|
|
9GLS
 
 | |
9GN8
 
 | | Crystal Structure of UFC1 E149D | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Kumar, M, Banerjee, S, Isupov, M.N, Wiener, R. | | Deposit date: | 2024-08-31 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | UFC1 reveals the multifactorial and plastic nature of oxyanion holes in E2 conjugating enzymes.
Nat Commun, 16, 2025
|
|
1MLI
 
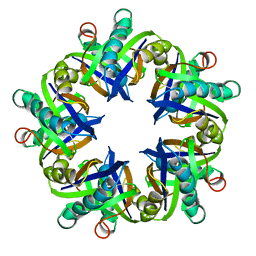 | |
1MMP
 
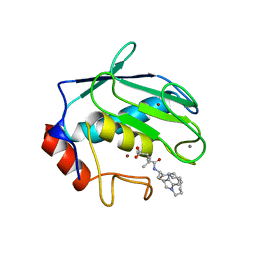 | | MATRILYSIN COMPLEXED WITH CARBOXYLATE INHIBITOR | | Descriptor: | 5-METHYL-3-(9-OXO-1,8-DIAZA-TRICYCLO[10.6.1.013,18]NONADECA-12(19),13,15,17-TETRAEN-10-YLCARBAMOYL)-HEXANOIC ACID, CALCIUM ION, GELATINASE A, ... | | Authors: | Browner, M.F, Smith, W.W, Castelhano, A.L. | | Deposit date: | 1995-03-22 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Matrilysin-inhibitor complexes: common themes among metalloproteases.
Biochemistry, 34, 1995
|
|
9GEZ
 
 | | Crystal structure of thioredoxin reductase from Cryptosporidium parvum in the "waiting" position | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase | | Authors: | Gabriele, F, Palerma, M, Ardini, M, Bogard, J, Chen, X.M, Williams, D.L, Angelucci, F. | | Deposit date: | 2024-08-08 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Targeting Apicomplexan Parasites: Structural and Functional Characterization of Cryptosporidium Thioredoxin Reductase as a Novel Drug Target.
Biochemistry, 64, 2025
|
|
1MC8
 
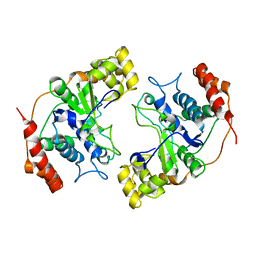 | | Crystal Structure of Flap Endonuclease-1 R42E mutant from Pyrococcus horikoshii | | Descriptor: | Flap Endonuclease-1 | | Authors: | Matsui, E, Musti, K.V, Abe, J, Yamazaki, K, Matsui, I, Harata, K. | | Deposit date: | 2002-08-06 | | Release date: | 2002-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Structure and Novel DNA Binding Sites Located in Loops of Flap Endonuclease-1 from Pyrococcus horikoshii
J.BIOL.CHEM., 277, 2002
|
|
9GIX
 
 | | Structure of the human mitochondrial pyruvate carrier in the apo-state | | Descriptor: | MBP-nanobody,Maltose/maltodextrin-binding periplasmic protein, Mitochondrial pyruvate carrier 1-like protein, Mitochondrial pyruvate carrier 2 | | Authors: | Sichrovsky, M, Lacabanne, D, Ruprecht, J.J, Rana, J.J, Stanik, K, Dionysopoulou, M, Sowton, A.P, King, M.S, Jones, S, Cooper, L, Hardwick, S.W, Paris, G, Chirgadze, D.Y, Ding, S, Fearnley, I.M, Palmer, S, Pardon, E, Steyaert, J, Leone, V, Forrest, L.R, Tavoulari, S, Kunji, E.R.S. | | Deposit date: | 2024-08-19 | | Release date: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Molecular basis of pyruvate transport and inhibition of the human mitochondrial pyruvate carrier.
Sci Adv, 11, 2025
|
|
7RGW
 
 | |
1MCL
 
 | | PRINCIPLES AND PITFALLS in DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), N-ACETYL-D-HIS-L-PRO-OH | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
9GSL
 
 | | Cryo-EM structure of human SLC35B1 in inward facing conformation | | Descriptor: | Fv-MBP, Solute carrier family 35 member B1 | | Authors: | Gulati, A, Ahn, D, Suades, A, Drew, D. | | Deposit date: | 2024-09-16 | | Release date: | 2025-05-21 | | Last modified: | 2025-08-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Stepwise ATP translocation into the endoplasmic reticulum by human SLC35B1.
Nature, 643, 2025
|
|
9G7G
 
 | | Structure of the clippase PaJOS from Pigmentiphaga aceris | | Descriptor: | PaJOS, Polyubiquitin-B, prop-2-en-1-amine | | Authors: | Baumann, U, Uthoff, M, Hermanns, T, Hofmann, K. | | Deposit date: | 2024-07-21 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A family of bacterial Josephin-like deubiquitinases with an irreversible cleavage mode.
Mol.Cell, 85, 2025
|
|
