9FXK
 
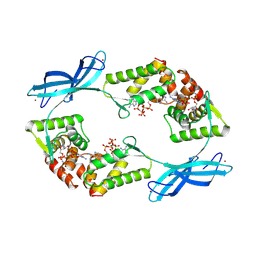 | |
1MG4
 
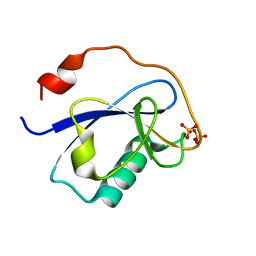 | | STRUCTURE OF N-TERMINAL DOUBLECORTIN DOMAIN FROM DCLK: WILD TYPE PROTEIN | | Descriptor: | DOUBLECORTIN-LIKE KINASE (N-TERMINAL DOMAIN), SULFATE ION | | Authors: | Kim, M.H, Cierpickil, T, Derewenda, U, Krowarsch, D, Feng, Y, Devedjiev, Y, Dauter, Z, Walsh, C.A, Otlewski, J, Bushweller, J.H, Derewenda, Z. | | Deposit date: | 2002-08-14 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | The DCX-domain Tandems of Doublecortin and Doublecortin-like Kinase
Nat.Struct.Biol., 10, 2003
|
|
5F0E
 
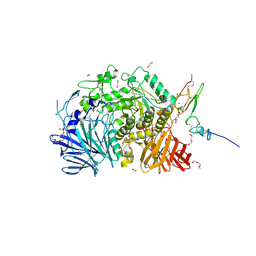 | | Murine endoplasmic reticulum alpha-glucosidase II | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2015-11-27 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7RUI
 
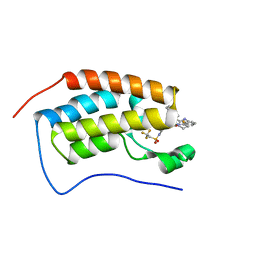 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with XR844 | | Descriptor: | Bromodomain-containing protein 4, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(5-{[(2-fluorophenyl)carbamoyl]amino}-1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-1H-indol-4-yl}-2,2,2-trifluoroethane-1-sulfonamide | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2021-08-17 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with XR844
To Be Published
|
|
9GMD
 
 | | MukEF in complex with the phage protein gp5.9 (focus) | | Descriptor: | Chromosome partition protein MukE, Chromosome partition protein MukF, Probable RecBCD inhibitor gp5.9 | | Authors: | Burmann, F, Wilkinson, O, Kimanius, D, Dillingham, M, Lowe, J. | | Deposit date: | 2024-08-28 | | Release date: | 2025-03-26 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanism of DNA capture by the MukBEF SMC complex and its inhibition by a viral DNA mimic.
Cell, 188, 2025
|
|
1MGQ
 
 | |
2P4W
 
 | | Crystal structure of heat shock regulator from Pyrococcus furiosus | | Descriptor: | SULFATE ION, Transcriptional regulatory protein arsR family | | Authors: | Liu, W, Vierke, G, Panjikar, S, Thomm, M, Ladenstein, R. | | Deposit date: | 2007-03-13 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Archaeal Heat Shock Regulator from Pyrococcus furiosus: A Molecular Chimera Representing Eukaryal and Bacterial Features.
J.Mol.Biol., 369, 2007
|
|
1M8R
 
 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 7.4) | | Descriptor: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase A2 | | Authors: | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | Deposit date: | 2002-07-25 | | Release date: | 2003-02-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
9G7D
 
 | | Crystal structure of ASGPR with bound IMP | | Descriptor: | Asialoglycoprotein receptor 1, CALCIUM ION, INOSINIC ACID | | Authors: | Schreuder, H.A, Hofmeister, A. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.588 Å) | | Cite: | Trivalent siRNA-Conjugates with Guanosine as ASGPR-Binder Show Potent Knock-Down In Vivo.
J.Med.Chem., 68, 2025
|
|
7RV4
 
 | |
9GB3
 
 | | Extended phiCD508 portal | | Descriptor: | gp45 - Portal protein | | Authors: | Wilson, J.S, Fagan, R.P, Bullough, P.A. | | Deposit date: | 2024-07-29 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Molecular mechanism of bacteriophage contraction structure of an S-layer-penetrating bacteriophage.
Life Sci Alliance, 8, 2025
|
|
1M9C
 
 | | X-ray crystal structure of Cyclophilin A/HIV-1 CA N-terminal domain (1-146) M-type Complex. | | Descriptor: | Cyclophilin A, HIV-1 Capsid | | Authors: | Howard, B.R, Vajdos, F.F, Li, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2002-07-28 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the catalytic mechanism of cyclophilin A
Nat.Struct.Biol., 10, 2003
|
|
7RVK
 
 | |
9GM6
 
 | |
7RWQ
 
 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one | | Descriptor: | 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
1MH3
 
 | | maltose binding-a1 homeodomain protein chimera, crystal form I | | Descriptor: | maltose binding-a1 homeodomain protein chimera | | Authors: | Ke, A, Wolberger, C. | | Deposit date: | 2002-08-19 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into binding cooperativity of MATa1/MATalpha2 from the crystal structure of a MATa1 homeodomain-maltose binding protein chimera
Protein Sci., 12, 2003
|
|
7RVC
 
 | |
9GB8
 
 | | Contracted phiCD508 tail | | Descriptor: | Phage tail sheath protein | | Authors: | Wilson, J.S, Fagan, R.P, Bullough, P.A. | | Deposit date: | 2024-07-29 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Molecular mechanism of bacteriophage contraction structure of an S-layer-penetrating bacteriophage.
Life Sci Alliance, 8, 2025
|
|
7RWF
 
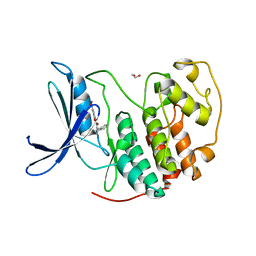 | | Crystal structure of CDK2 in complex with TW8672 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
9G0W
 
 | | auxin transporter PIN8 as asymmetric dimer (inward/outward) with 2,4-D bound in the inward vestibule prebinding state | | Descriptor: | (2,4-DICHLOROPHENOXY)ACETIC ACID, 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Auxin efflux carrier component 8, ... | | Authors: | Ung, K.L, Stokes, D.L, Pedersen, B.P. | | Deposit date: | 2024-07-09 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Transport of herbicides by PIN-FORMED auxin transporters.
Biorxiv, 2024
|
|
1MHX
 
 | | Crystal Structures of the redesigned protein G variant NuG1 | | Descriptor: | immunoglobulin-binding protein G | | Authors: | Nauli, S, Kuhlman, B, Le Trong, I, Stenkamp, R.E, Teller, D.C, Baker, D. | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and increased stabilization of the protein G variants with switched folding pathways NuG1 and NuG2
Protein Sci., 11, 2002
|
|
7RV7
 
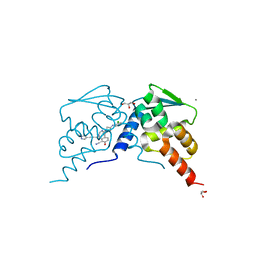 | |
5F1C
 
 | | Crystal structure of an invertebrate P2X receptor from the Gulf Coast tick in the presence of ATP and Zn2+ ion at 2.9 Angstroms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Putative uncharacterized protein, ... | | Authors: | Kasuya, G, Hattori, M, Ishitani, R, Nureki, O. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insights into Divalent Cation Modulations of ATP-Gated P2X Receptor Channels
Cell Rep, 14, 2016
|
|
9G10
 
 | | auxin transporter PIN8 as asymmetric dimer (outward/inward) with 4-CPA bound in the outward partly released state | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(4-chloranylphenoxy)ethanoic acid, Auxin efflux carrier component 8, ... | | Authors: | Ung, K.L, Stokes, D.L, Pedersen, B.P. | | Deposit date: | 2024-07-09 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Transport of herbicides by PIN-FORMED auxin transporters.
Biorxiv, 2024
|
|
7RVF
 
 | |
