7R8S
 
 | |
1LVK
 
 | | X-RAY CRYSTAL STRUCTURE OF THE MG (DOT) 2'(3')-O-(N-METHYLANTHRANILOYL) NUCLEOTIDE BOUND TO DICTYOSTELIUM DISCOIDEUM MYOSIN MOTOR DOMAIN | | Descriptor: | 2'(3')-O-N-METHYLANTHRANILOYL-ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Bauer, C.B, Kuhlman, P.A, Bagshaw, C.R, Rayment, I. | | Deposit date: | 1997-09-05 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystal structure and solution fluorescence characterization of Mg.2'(3')-O-(N-methylanthraniloyl) nucleotides bound to the Dictyostelium discoideum myosin motor domain.
J.Mol.Biol., 274, 1997
|
|
1MAI
 
 | | STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM PHOSPHOLIPASE C DELTA IN COMPLEX WITH INOSITOL TRISPHOSPHATE | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PHOSPHOLIPASE C DELTA-1 | | Authors: | Ferguson, K.M, Lemmon, M.A, Schlessinger, J, Sigler, P.B. | | Deposit date: | 1996-05-23 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain.
Cell(Cambridge,Mass.), 83, 1995
|
|
7R7V
 
 | | Crystal structure of HLA-B*5301 complex with an HIV-1 Gag-derived epitope QW9 | | Descriptor: | Beta-2-microglobulin, GLN-ALA-SER-GLN-GLU-VAL-LYS-ASN-TRP, GLYCEROL, ... | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
9GMA
 
 | | MukBEF in a DNA capture state (dimer) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, Chromosome partition protein MukB, ... | | Authors: | Burmann, F, Lowe, J. | | Deposit date: | 2024-08-28 | | Release date: | 2025-03-26 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Mechanism of DNA capture by the MukBEF SMC complex and its inhibition by a viral DNA mimic.
Cell, 188, 2025
|
|
7RRE
 
 | | Carbonic Anhydrase II in complex with Beta-Galactose-2C | | Descriptor: | 2-[1-(1,1,3-trioxo-2,3-dihydro-1H-1lambda~6~,2-benzothiazol-6-yl)-1H-1,2,3-triazol-4-yl]ethyl beta-L-gulopyranoside, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | McKenna, R, Combs, J.E. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Inhibition of Carbonic Anhydrase IX and Monocarboxylate Transporters 1 and 4 in breast cancer via novel inhibition with Beta-Galactose 2C
To Be Published
|
|
9GB5
 
 | | Contracted phiCD508 neck | | Descriptor: | gp45 - Portal protein, gp50 - Portal adaptor protein, gp51 - Neck valve protein, ... | | Authors: | Wilson, J.S, Fagan, R.P, Bullough, P.A. | | Deposit date: | 2024-07-29 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Molecular mechanism of bacteriophage contraction structure of an S-layer-penetrating bacteriophage.
Life Sci Alliance, 8, 2025
|
|
9GM9
 
 | | MukBEF in a DNA capture state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, Chromosome partition protein MukB, ... | | Authors: | Burmann, F, Lowe, J. | | Deposit date: | 2024-08-28 | | Release date: | 2025-03-26 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Mechanism of DNA capture by the MukBEF SMC complex and its inhibition by a viral DNA mimic.
Cell, 188, 2025
|
|
1LVB
 
 | | CATALYTICALLY INACTIVE TOBACCO ETCH VIRUS PROTEASE COMPLEXED WITH SUBSTRATE | | Descriptor: | CATALYTIC DOMAIN OF THE NUCLEAR INCLUSION PROTEIN A (NIA), GLYCEROL, OLIGOPEPTIDE SUBSTRATE FOR THE PROTEASE | | Authors: | Phan, J, Zdanov, A, Evdokimov, A.G, Tropea, J.E, Peters III, H.K, Kapust, R.B, Li, M, Wlodawer, A, Waugh, D.S. | | Deposit date: | 2002-05-28 | | Release date: | 2002-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the substrate specificity of tobacco etch virus protease.
J.Biol.Chem., 277, 2002
|
|
9FZF
 
 | | Transcriptional repressor NrdR from E. coli, ADP/dATP bound state | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bimai, O, Rozman Grinberg, I, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2024-07-05 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Bacterial transcriptional repressor NrdR - a flexible multifactorial nucleotide sensor.
Febs J., 292, 2025
|
|
1LVQ
 
 | | IC3 of CB1 Bound to G(alpha)i | | Descriptor: | Cannabinoid receptor 1 | | Authors: | Ulfers, A.L, McMurry, J.L, Miller, A, Wang, L, Kendall, D.A, Mierke, D.F. | | Deposit date: | 2002-05-29 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Cannabinoid receptor-G protein interactions: G(alphai1)-bound structures of IC3 and a mutant with altered G protein specificity.
Protein Sci., 11, 2002
|
|
9GAY
 
 | | Extended phiCD508 capsid | | Descriptor: | gp48 - Minor capsid protein, gp49 - Major capsid protein | | Authors: | Wilson, J.S, Fagan, R.P, Bullough, P.A. | | Deposit date: | 2024-07-29 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Molecular mechanism of bacteriophage contraction structure of an S-layer-penetrating bacteriophage.
Life Sci Alliance, 8, 2025
|
|
1LVS
 
 | | THE SOLUTION STRUCTURE OF D(G4T4G3)2 | | Descriptor: | 5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*G)-3' | | Authors: | Crnugelj, M, Hud, N.V, Plavec, J. | | Deposit date: | 2002-05-29 | | Release date: | 2002-07-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of d(G(4)T(4)G(3))(2): a bimolecular G-quadruplex with a novel fold.
J.Mol.Biol., 320, 2002
|
|
1LW9
 
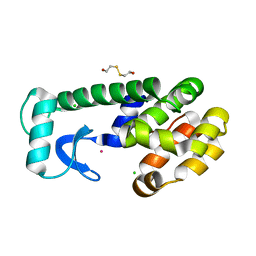 | | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME, ... | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H.M, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-05-30 | | Release date: | 2003-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability.
Biophys.Chem., 100, 2003
|
|
7RVH
 
 | |
9GB1
 
 | | Extended phiCD508 tail | | Descriptor: | gp55 - Tail sheath protein, gp56 - Tail tube protein | | Authors: | Wilson, J.S, Fagan, R.P, Bullough, P.A. | | Deposit date: | 2024-07-29 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Molecular mechanism of bacteriophage contraction structure of an S-layer-penetrating bacteriophage.
Life Sci Alliance, 8, 2025
|
|
7RWE
 
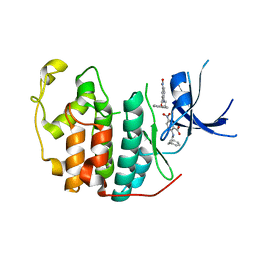 | | Crystal structure of CDK2 liganded with compound GPHR787 | | Descriptor: | 5-nitro-2-[(3-phenylpropyl)amino]benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
1LWC
 
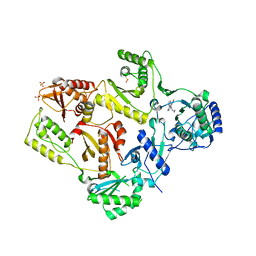 | | CRYSTAL STRUCTURE OF M184V MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
1LWY
 
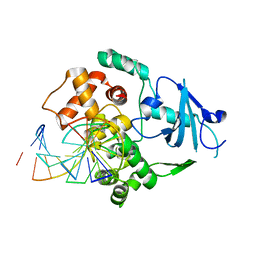 | | hOgg1 Borohydride-Trapped Intermediate without 8-oxoguanine | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(PED)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 8-OXOGUANINE DNA GLYCOSYLASE | | Authors: | Fromme, J.C, Bruner, S.D, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2002-06-03 | | Release date: | 2003-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Product-Assisted Catalysis in Base Excision DNA Repair
Nat.Struct.Biol., 10, 2003
|
|
7R7W
 
 | | Crystal structure of HLA-B*5301 complex with an HIV-1 Gag-derived epitope QW9 S3T variant | | Descriptor: | Beta-2-microglobulin, GLN-ALA-THR-GLN-GLU-VAL-LYS-ASN-TRP, MHC class I antigen | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
9GB7
 
 | | Extended phiCD508 neck | | Descriptor: | gp45 - Portal protein, gp50 - Portal adaptor protein, gp51 - Neck valve protein, ... | | Authors: | Wilson, J.S, Fagan, R.P, Bullough, P.A. | | Deposit date: | 2024-07-29 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular mechanism of bacteriophage contraction structure of an S-layer-penetrating bacteriophage.
Life Sci Alliance, 8, 2025
|
|
1LXF
 
 | | Structure of the Regulatory N-domain of Human Cardiac Troponin C in Complex with Human Cardiac Troponin-I(147-163) and Bepridil | | Descriptor: | 1-ISOBUTOXY-2-PYRROLIDINO-3[N-BENZYLANILINO] PROPANE, CALCIUM ION, TROPONIN C, ... | | Authors: | Wang, X, Li, M.X, Sykes, B.D. | | Deposit date: | 2002-06-05 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the regulatory N-domain of human
cardiac troponin C in complex with human cardiac
troponin I147-163 and bepridil.
J.Biol.Chem., 277, 2002
|
|
7RVD
 
 | |
1M06
 
 | | Structural Studies of Bacteriophage alpha3 Assembly, X-Ray Crystallography | | Descriptor: | 5'-D(P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR)P*(3DR))-3', Capsid Protein, Major spike protein, ... | | Authors: | Bernal, R.A, Hafenstein, S, Olson, N.H, Bowman, V, Chipman, P.R, Baker, T.S, Fane, B.A, Rossmann, M.G. | | Deposit date: | 2002-06-12 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Studies of Bacteriophage alpha3 Assembly
J.Mol.Biol., 325, 2003
|
|
7RWN
 
 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-5-{4-[(dimethylamino)methyl]anilino}-2-methylpyridazin-3(2H)-one | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-5-{4-[(dimethylamino)methyl]anilino}-2-methylpyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
