3MO2
 
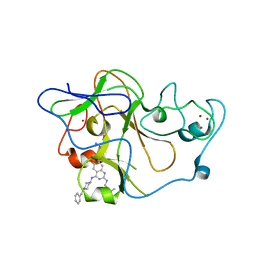 | | human G9a-like (GLP, also known as EHMT1) in complex with inhibitor E67 | | Descriptor: | 7-[(5-aminopentyl)oxy]-N~4~-(1-benzylpiperidin-4-yl)-N~2~-[3-(dimethylamino)propyl]-6-methoxyquinazoline-2,4-diamine, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, ... | | Authors: | Chang, Y, Horton, J.R, Cheng, X. | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases.
J.Mol.Biol., 400, 2010
|
|
1O2G
 
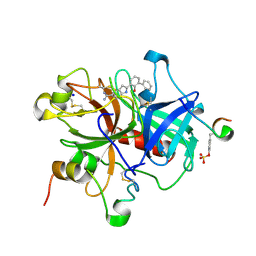 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 3-{5-[AMINO(IMINIO)METHYL]-1H-INDOL-2-YL}-1,1'-BIPHENYL-2-OLATE, ACETYL HIRUDIN, SODIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, W.F, Hui, H, Breitenbucher, G, Allen, D, Janc, J. | | Deposit date: | 2003-03-06 | | Release date: | 2003-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating
Binding of Active Site-Directed Serine Protease Inhibitors
J.Mol.Biol., 329, 2003
|
|
2W15
 
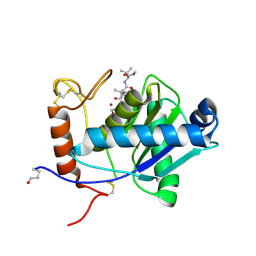 | | High-resolution crystal structure of the P-I snake venom metalloproteinase BaP1 in complex with a peptidomimetic: insights into inhibitor binding | | Descriptor: | (2R,3R)-N^1^-[(1S)-2,2-DIMETHYL-1-(METHYLCARBAMOYL)PROPYL]-N^4^-HYDROXY-2-(2-METHYLPROPYL)-3-{[(1,3-THIAZOL-2-YLCARBONYL)AMINO]METHYL}BUTANEDIAMIDE, GLYCEROL, ZINC ION, ... | | Authors: | Lingott, T.J, Schleberger, C, Gutierrez, J.M, Merfort, I. | | Deposit date: | 2008-10-14 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | High-Resolution Crystal Structure of the Snake Venom Metalloproteinase Bap1 Complexed with a Peptidomimetic: Insight Into Inhibitor Binding.
Biochemistry, 48, 2009
|
|
4QVL
 
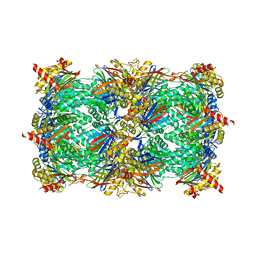 | | yCP in complex with bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-15 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4YQX
 
 | | Mouse IL-2 Bound to JES6-1 scFv Fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-2, JES6-1 VH domain, ... | | Authors: | Spangler, J.B, Luca, V.C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2015-03-13 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.826 Å) | | Cite: | Antibodies to Interleukin-2 Elicit Selective T Cell Subset Potentiation through Distinct Conformational Mechanisms.
Immunity, 42, 2015
|
|
4QSS
 
 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with N-Methyl-2-pyrrolidone (NMP) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-methylpyrrolidin-2-one, ATPase family AAA domain-containing protein 2, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
6AYO
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with 5'-deoxy-5'-Propyl-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-propylpyrrolidin-3-ol, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
1O47
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU82209. | | Descriptor: | N-ACETYL-N-[1-(1,1'-BIPHENYL-4-YLMETHYL)-2-OXOAZEPAN-3-YL]-4-[DIFLUORO(PHOSPHONO)METHYL]PHENYLALANINAMIDE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
3QHT
 
 | | Crystal Structure of the Monobody ySMB-1 bound to yeast SUMO | | Descriptor: | GLYCEROL, Monobody ySMB-1, Ubiquitin-like protein SMT3 | | Authors: | Koide, S, Gilbreth, R.N. | | Deposit date: | 2011-01-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Isoform-specific monobody inhibitors of small ubiquitin-related modifiers engineered using structure-guided library design.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2VTS
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | 5-[(4-AMINOCYCLOHEXYL)AMINO]-7-(PROPAN-2-YLAMINO)PYRAZOLO[1,5-A]PYRIMIDINE-3-CARBONITRILE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
4M3M
 
 | |
4DEI
 
 | | Crystal structure of c-Met in complex with triazolopyridinone inhibitor 24 | | Descriptor: | 3-{(1S)-1-[3-(2-methoxyethoxy)quinolin-6-yl]ethyl}-5-(3-methyl-1,2-thiazol-5-yl)-3,5-dihydro-4H-[1,2,3]triazolo[4,5-c]pyridin-4-one, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2012-01-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and optimization of a potent and selective triazolopyridinone series of c-Met inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6U11
 
 | | Xenopus laevis N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (NAGPA) (C46S C219S C453S C480S C486S) with CTD mostly flexible | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Mannose-6-Phosphate Uncovering Enzyme.
Structure, 28, 2020
|
|
1O9R
 
 | | The X-ray crystal structure of Agrobacterium tumefaciens Dps, a member of the family that protect DNA without binding | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AGROBACTERIUM TUMEFACIENS DPS, ... | | Authors: | Ilari, A, Ceci, P, Chiancone, E. | | Deposit date: | 2002-12-18 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Dps Protein of Agrobacterium Tumefaciens Does not Bind to DNA But Protects It Toward Oxidative Cleavage: X-Ray Crystal Structure, Iron Binding, and Hydroxyl-Radical Scavenging Properties
J.Biol.Chem., 278, 2003
|
|
5RA2
 
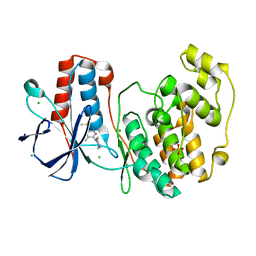 | |
1S5H
 
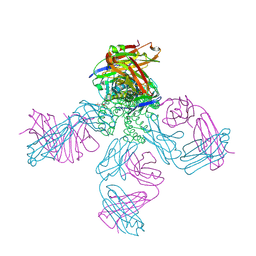 | | Potassium Channel Kcsa-Fab Complex T75C mutant in K+ | | Descriptor: | ANTIBODY FAB FRAGMENT HEAVY CHAIN, ANTIBODY FAB FRAGMENT LIGHT CHAIN, DIACYL GLYCEROL, ... | | Authors: | Mackinnon, R, Zhou, M. | | Deposit date: | 2004-01-20 | | Release date: | 2004-05-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A mutant KcsA K(+) channel with altered conduction properties and selectivity filter ion distribution.
J.Mol.Biol., 338, 2004
|
|
1NNY
 
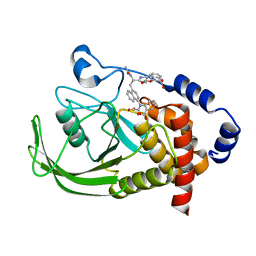 | | Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor Compound 23 Using a Linked-Fragment Strategy | | Descriptor: | 3-({5-[(N-ACETYL-3-{4-[(CARBOXYCARBONYL)(2-CARBOXYPHENYL)AMINO]-1-NAPHTHYL}-L-ALANYL)AMINO]PENTYL}OXY)-2-NAPHTHOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Pei, Z, Xin, Z, Lubben, T, Trevillyan, J.M, Stashko, M.A, Ballaron, S.J, Liang, H, Huang, F, Hutchins, C.W, Fesik, S.W, Jirousek, M.R. | | Deposit date: | 2003-01-14 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor
Using a Linked-Fragment Strategy
J.Am.Chem.Soc., 125, 2003
|
|
5RC0
 
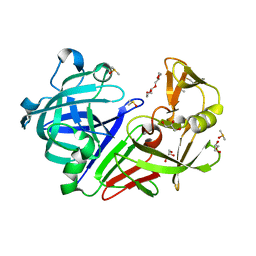 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library E06a | | Descriptor: | (3-endo)-8-benzyl-8-azabicyclo[3.2.1]octan-3-ol, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
4R4I
 
 | | Structure of RPA70N in complex with 5-(4-((6-(5-carboxyfuran-2-yl)-1-thioxo-3,4-dihydroisoquinolin-2(1H)-yl)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-(4-{[6-(5-carboxyfuran-2-yl)-1-thioxo-3,4-dihydroisoquinolin-2(1H)-yl]methyl}phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
8IDQ
 
 | | Crystal structure of reducing-end xylose-releasing exoxylanase in GH30 from Talaromyces cellulolyticus with xylose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nakamichi, Y, Watanabe, M, Fujii, T, Inoue, H, Morita, T. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of reducing-end xylose-releasing exoxylanase in subfamily 7 of glycoside hydrolase family 30.
Proteins, 91, 2023
|
|
4M22
 
 | | Crystal Structure of small molecule acrylamide 16 covalently bound to K-Ras G12C | | Descriptor: | 1-{4-[(2,4-dichlorophenoxy)acetyl]piperazin-1-yl}propan-1-one, GUANOSINE-5'-DIPHOSPHATE, K-Ras GTPase, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
6K0A
 
 | | cryo-EM structure of an archaeal Ribonuclease P | | Descriptor: | 50S ribosomal protein L7Ae, RPR, Ribonuclease P protein component 1, ... | | Authors: | Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-19 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-electron microscopy structure of an archaeal ribonuclease P holoenzyme.
Nat Commun, 10, 2019
|
|
6DYW
 
 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with (3R,4S)-1-((4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl)-4-(((3-(1-benzyl-1H-1,2,3-triazol-4-yl)propyl)thio)methyl)pyrrolidin-3-ol | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-({[3-(1-benzyl-1H-1,2,3-triazol-4-yl)propyl]sulfanyl}methyl)pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ... | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2018-07-02 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Selective Inhibitors of Helicobacter pylori Methylthioadenosine Nucleosidase and Human Methylthioadenosine Phosphorylase.
J. Med. Chem., 62, 2019
|
|
8IDP
 
 | | Crystal structure of reducing-end xylose-releasing exoxylanase in GH30 from Talaromyces cellulolyticus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamichi, Y, Watanabe, M, Fujii, T, Inoue, H, Morita, T. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of reducing-end xylose-releasing exoxylanase in subfamily 7 of glycoside hydrolase family 30.
Proteins, 91, 2023
|
|
5RFB
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1271660837 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1-methyl-1H-1,2,3-triazol-4-yl)methyl]ethanamine | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
