1MDF
 
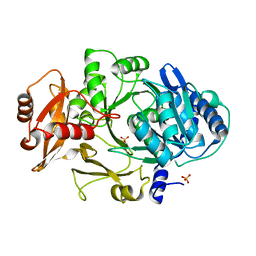 | | CRYSTAL STRUCTURE OF DhbE IN ABSENCE OF SUBSTRATE | | Descriptor: | 2,3-dihydroxybenzoate-AMP ligase, SULFATE ION | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6DR8
 
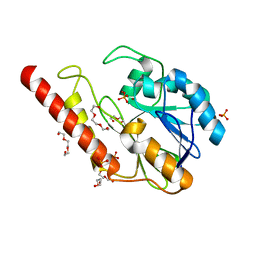 | | Metallo-beta-lactamase from Cronobacter sakazakii (Enterobacter sakazakii) HARLDQ motif mutant S60/R118H/Q121H/K254H | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Monteiro Pedroso, M, Waite, D, Natasa, M, McGeary, R, Guddat, L, Hugenholtz, P, Schenk, G. | | Deposit date: | 2018-06-11 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.476 Å) | | Cite: | Broad spectrum antibiotic-degrading metallo-beta-lactamases are phylogenetically diverse
Protein Cell, 2020
|
|
3V49
 
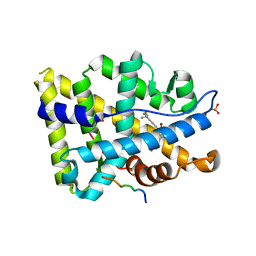 | | Structure of ar lbd with activator peptide and sarm inhibitor 1 | | Descriptor: | 4-[(4R)-4-(4-hydroxyphenyl)-3,4-dimethyl-2,5-dioxoimidazolidin-1-yl]-2-(trifluoromethyl)benzonitrile, Androgen receptor, activator peptide, ... | | Authors: | Nique, F, Hebbe, S, Peixoto, C, Annoot, D, Lefrancois, J.-M, Duval, E, Michoux, L, Triballeau, N, Lemoullec, J.-M, Mollat, P, Thauvin, M, Prange, T, Minet, D, Clement-Lacroix, P, Robin-Jagerschmidt, C, Fleury, D, Guedin, D, Deprez, P. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of diarylhydantoins as new selective androgen receptor modulators.
J.Med.Chem., 55, 2012
|
|
1M82
 
 | | SOLUTION STRUCTURE OF THE COMPLEMENTARY RNA PROMOTER OF INFLUENZA A VIRUS | | Descriptor: | RNA (25-MER): THE COMPLEMENTARY RNA PROMOTER OF INFLUENZA A VIRUS | | Authors: | Park, C.-J, Bae, S.-H, Lee, M.-K, Varani, G, Choi, B.-S. | | Deposit date: | 2002-07-24 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the influenza A virus cRNA promoter: implications for differential recognition of viral promoter structures by RNA-dependent RNA polymerase
NUCLEIC ACIDS RES., 31, 2003
|
|
1MDB
 
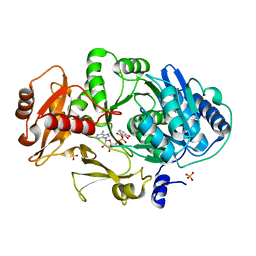 | | CRYSTAL STRUCTURE OF DhbE IN COMPLEX WITH DHB-ADENYLATE | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2,3-dihydroxybenzoate-AMP ligase, ADENOSINE MONOPHOSPHATE, ... | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6E8A
 
 | | Crystal structure of DcrB from Salmonella enterica at 1.92 Angstroms resolution | | Descriptor: | DUF1795 domain-containing protein | | Authors: | Rasmussen, D.M, Soens, R.W, Bhattacharyya, B, May, J.F. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The structure of DcrB, a lipoprotein from Salmonella enterica, reveals flexibility in the N-terminal segment of the Mog1p/PsbP-like fold.
J. Struct. Biol., 204, 2018
|
|
3NV2
 
 | |
6EN6
 
 | | Crystal structure B of the Angiotensin-1 converting enzyme N-domain in complex with a diprolyl inhibitor. | | Descriptor: | (2~{S})-1-[(2~{S})-2-[[(1~{S})-1-[(2~{S})-1-[(2~{S})-2-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]pyrrolidin-2-yl]-2-oxidanyl-2-oxidanylidene-ethyl]amino]propanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R, Fienberg, S, Chibale, K, Sturrock, E.D. | | Deposit date: | 2017-10-04 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Design and Development of a Potent and Selective Novel Diprolyl Derivative That Binds to the N-Domain of Angiotensin-I Converting Enzyme.
J. Med. Chem., 61, 2018
|
|
1JYD
 
 | | Crystal Structure of Recombinant Human Serum Retinol-Binding Protein at 1.7 A Resolution | | Descriptor: | GLYCEROL, PLASMA RETINOL-BINDING PROTEIN | | Authors: | Greene, L.H, Chrysina, E.D, Irons, L.I, Papageorgiou, A.C, Acharya, K.R, Brew, K. | | Deposit date: | 2001-09-12 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of conserved residues in structure and stability: Tryptophans of human serum
retinol-binding protein, a model for the lipocalin superfamily
Protein Sci., 10, 2001
|
|
4BJL
 
 | |
6EN5
 
 | | Crystal structure A of the Angiotensin-1 converting enzyme N-domain in complex with a diprolyl inhibitor. | | Descriptor: | (2~{S})-1-[(2~{S})-2-[[(1~{S})-1-[(2~{S})-1-[(2~{S})-2-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]pyrrolidin-2-yl]-2-oxidanyl-2-oxidanylidene-ethyl]amino]propanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R, Fienberg, S, Chibale, K, Sturrock, E.D. | | Deposit date: | 2017-10-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Design and Development of a Potent and Selective Novel Diprolyl Derivative That Binds to the N-Domain of Angiotensin-I Converting Enzyme.
J. Med. Chem., 61, 2018
|
|
6EZ4
 
 | | NMR structure of the C-terminal domain of the human RPAP3 protein | | Descriptor: | RNA polymerase II-associated protein 3 | | Authors: | Fabre, P, Chagot, M.E, Bragantini, B, Manival, X, Quinternet, M. | | Deposit date: | 2017-11-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | The RPAP3-Cterminal domain identifies R2TP-like quaternary chaperones.
Nat Commun, 9, 2018
|
|
2DGY
 
 | | Solution structure of the eukaryotic initiation factor 1A in MGC11102 protein | | Descriptor: | MGC11102 protein | | Authors: | Abe, C, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-16 | | Release date: | 2006-09-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the eukaryotic initiation factor 1A in MGC11102 protein
To be Published
|
|
3R1W
 
 | | Crystal structure of a carbonic anhydrase from a crude oil degrading psychrophilic library | | Descriptor: | carbonic anhydrase | | Authors: | Petit, P, Xu, X, Cui, H, Brown, G, Dong, A, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2011-03-11 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of a carbonic anhydrase from a crude oil degrading psychrophilic library
To be Published
|
|
1LUL
 
 | | DB58, A LEGUME LECTIN FROM DOLICHOS BIFLORUS | | Descriptor: | CALCIUM ION, LECTIN DB58, MANGANESE (II) ION | | Authors: | Hamelryck, T.W, Bouckaert, J, Dao-Thi, M.H, Wyns, L, Etzler, M, Loris, R. | | Deposit date: | 1998-06-30 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus.
J.Mol.Biol., 286, 1999
|
|
6DHM
 
 | | Bovine glutamate dehydrogenase complexed with zinc | | Descriptor: | GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | Authors: | Smith, T.J. | | Deposit date: | 2018-05-20 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A novel mechanism of V-type zinc inhibition of glutamate dehydrogenase results from disruption of subunit interactions necessary for efficient catalysis.
FEBS J., 278, 2011
|
|
4OP2
 
 | | GKRP bound to AMG-0471 and Sorbitol-6-Phosphate | | Descriptor: | (2S)-2-{4'-[(6-aminopyridin-3-yl)sulfonyl]biphenyl-4-yl}-1,1,1-trifluoropropan-2-ol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | Jordan, S.R, Chmait, S. | | Deposit date: | 2014-02-04 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Small Molecule Disruptors of the Glucokinase-Glucokinase Regulatory Protein Interaction: 5. A Novel Aryl Sulfone Series, Optimization Through Conformational Analysis.
J.Med.Chem., 58, 2015
|
|
2AMY
 
 | | X-Ray Structure of Human Phosphomannomutase 2 (PMM2) | | Descriptor: | 1,2-ETHANEDIOL, GLYCINE, Phosphomannomutase 2 | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, McCoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-08-10 | | Release date: | 2005-08-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | X-Ray Structure of Human Phosphomannomutase 2 (PMM2)
To be Published
|
|
4O88
 
 | | Crystal structure of a C-tagged Nuclease | | Descriptor: | CHLORIDE ION, N-tagged Nuclease, SULFATE ION | | Authors: | Chakravarty, A.K, Smith, P, Shuman, S. | | Deposit date: | 2013-12-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure, mechanism, and specificity of a eukaryal tRNA restriction enzyme involved in self-nonself discrimination.
Cell Rep, 7, 2014
|
|
4MPK
 
 | | Crystal structure of the complex of buffalo signaling protein SPB-40 with N-acetylglucosamine at 2.65 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, GLYCEROL | | Authors: | Yamini, S, Chaudhary, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-09-13 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the complex of buffalo signaling protein SPB-40 with N-acetylglucosamine at 2.65 A resolution
To be Published
|
|
2VUV
 
 | | Crystal structure of Codakine at 1.3A resolution | | Descriptor: | CALCIUM ION, CITRIC ACID, CODAKINE, ... | | Authors: | Gourdine, J.P, Cioci, G.C, Miguet, L, Unverzagt, C, Varrot, A, Gauthier, C, Smith-Ravin, E.J, Imberty, A. | | Deposit date: | 2008-05-30 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High Affinity Interaction between a Bivalve C-Type Lectin and a Biantennary Complex-Type N-Glycan Revealed by Crystallography and Microcalorimetry.
J.Biol.Chem., 283, 2008
|
|
6E0S
 
 | | Crystal structure of MEM-A1, a subclass B3 metallo-beta-lactamase isolated from a soil metagenome library | | Descriptor: | GLYCEROL, MEM-A1, PHOSPHATE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Lau, C, Topp, E, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-06 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of MEM-A1, a subclass B3 metallo-beta-lactamase isolated from a soil metagenome library
To Be Published
|
|
6F8W
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-18a | | Descriptor: | 3-[3-(3-cyclopentyloxy-4-methoxy-phenyl)pyrazol-1-yl]-1-morpholin-4-yl-propan-1-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6F9V
 
 | |
4P73
 
 | | PheRS in complex with compound 1a | | Descriptor: | 1-{3-[(4-pyridin-2-ylpiperazin-1-yl)sulfonyl]phenyl}-3-(1,3-thiazol-2-yl)urea, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-03-25 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | The role of a novel auxiliary pocket in bacterial phenylalanyl-tRNA synthetase druggability.
J.Biol.Chem., 289, 2014
|
|
